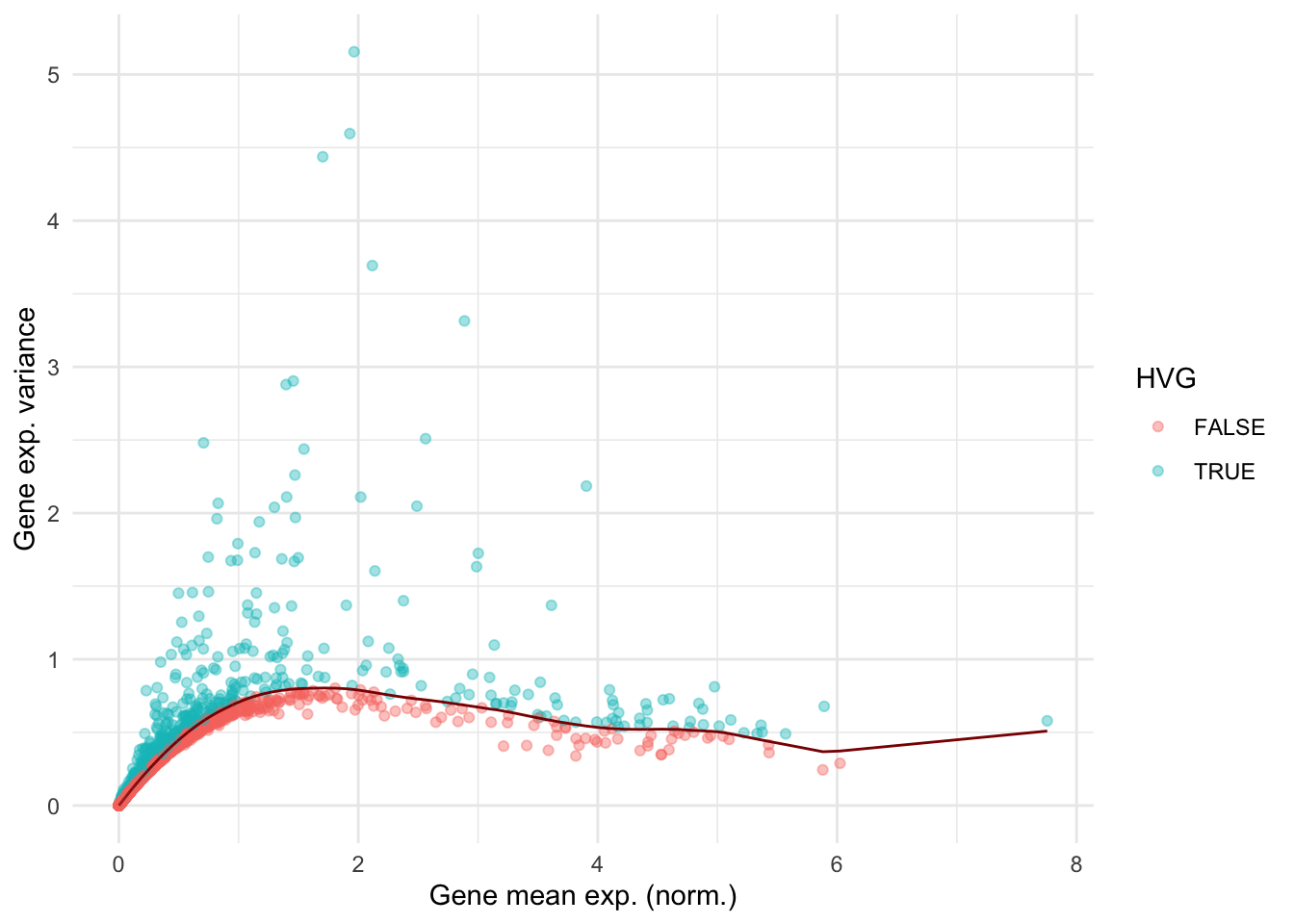
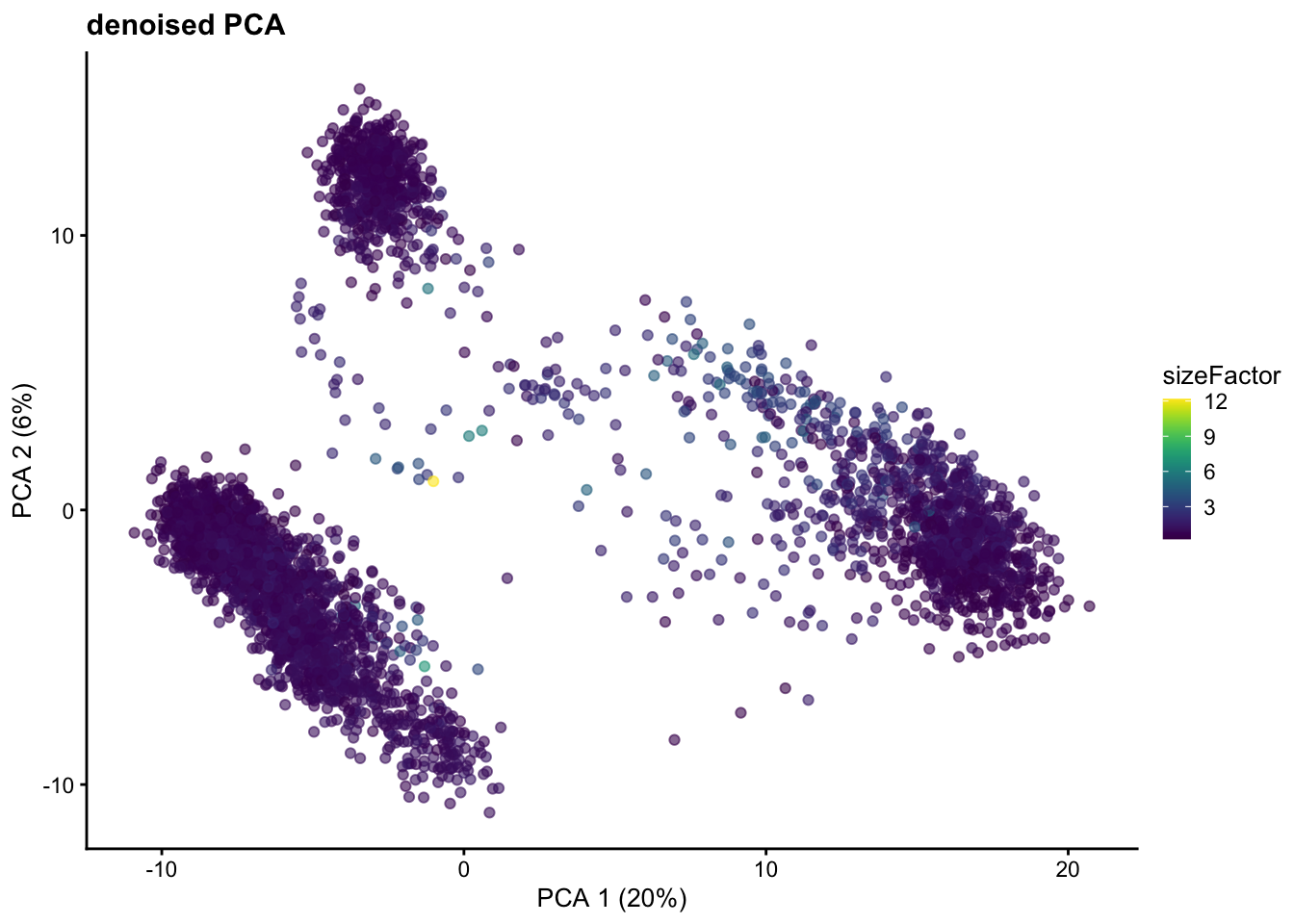
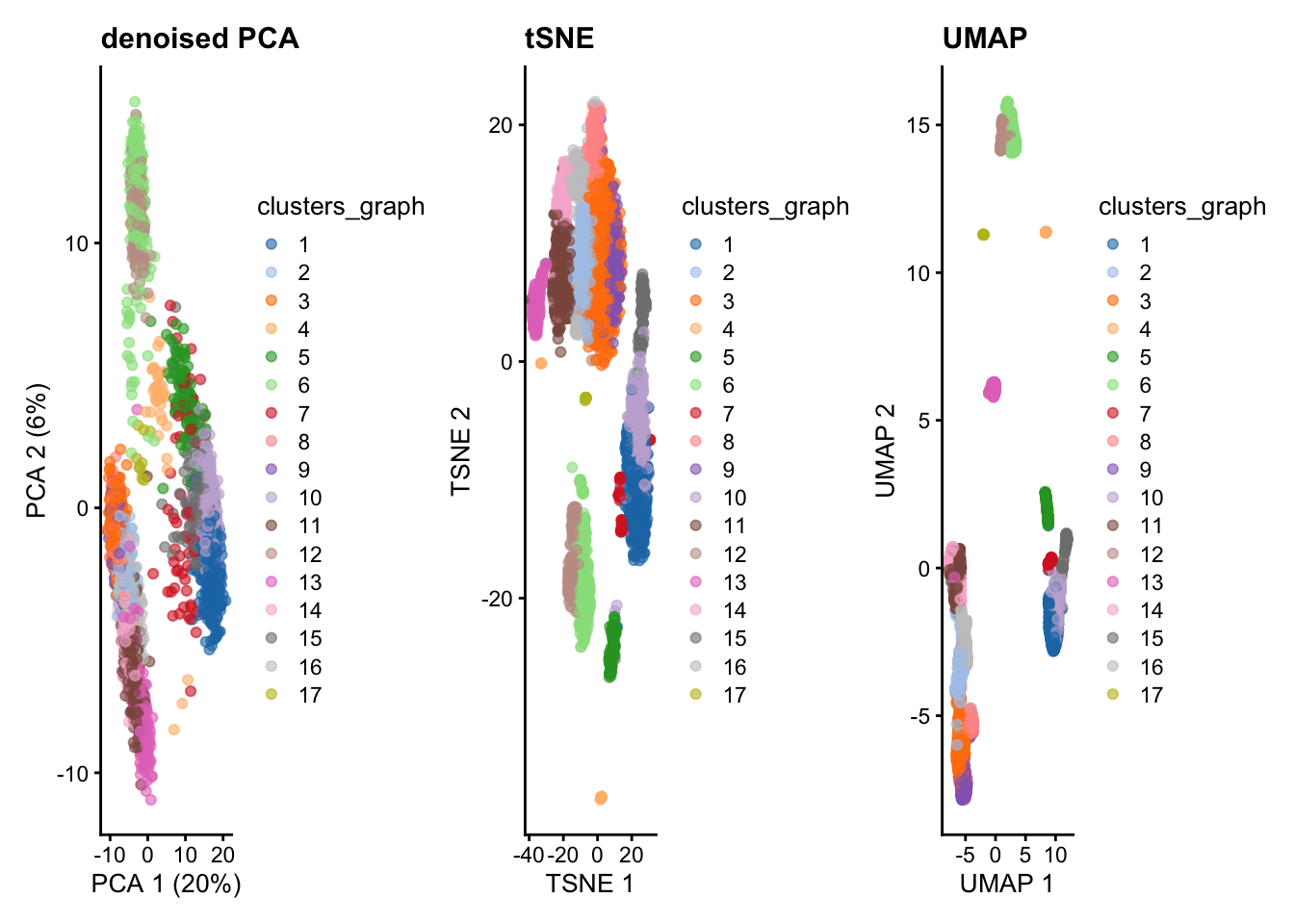
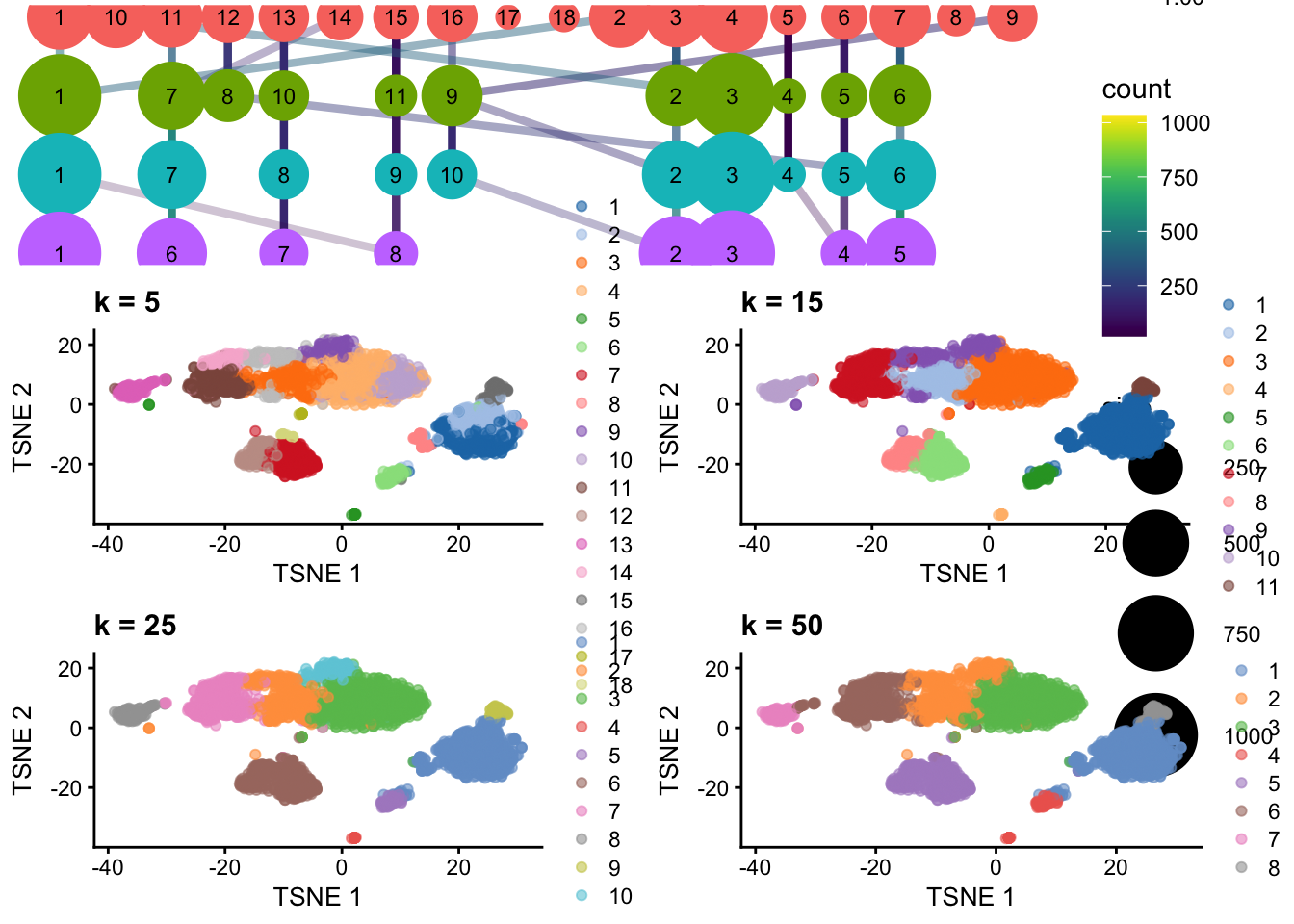
── Attaching core tidyverse packages ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(SingleCellExperiment)Loading required package: SummarizedExperiment
Loading required package: MatrixGenerics
Loading required package: matrixStats
Attaching package: 'matrixStats'
The following object is masked from 'package:dplyr':
count
Attaching package: 'MatrixGenerics'
The following objects are masked from 'package:matrixStats':
colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse, colCounts, colCummaxs, colCummins, colCumprods, colCumsums, colDiffs, colIQRDiffs, colIQRs, colLogSumExps,
colMadDiffs, colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats, colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds, colSums2, colTabulates,
colVarDiffs, colVars, colWeightedMads, colWeightedMeans, colWeightedMedians, colWeightedSds, colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet, rowCollapse,
rowCounts, rowCummaxs, rowCummins, rowCumprods, rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps, rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks, rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars, rowWeightedMads, rowWeightedMeans,
rowWeightedMedians, rowWeightedSds, rowWeightedVars
Loading required package: GenomicRanges
Loading required package: stats4
Loading required package: BiocGenerics
Attaching package: 'BiocGenerics'
The following objects are masked from 'package:lubridate':
intersect, setdiff, union
The following objects are masked from 'package:dplyr':
combine, intersect, setdiff, union
The following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabs
The following objects are masked from 'package:base':
anyDuplicated, aperm, append, as.data.frame, basename, cbind, colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep, grepl, intersect, is.unsorted,
lapply, Map, mapply, match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank, rbind, Reduce, rownames, sapply, setdiff, table, tapply, union, unique,
unsplit, which.max, which.min
Loading required package: S4Vectors
Attaching package: 'S4Vectors'
The following objects are masked from 'package:lubridate':
second, second<-
The following objects are masked from 'package:dplyr':
first, rename
The following object is masked from 'package:tidyr':
expand
The following object is masked from 'package:utils':
findMatches
The following objects are masked from 'package:base':
expand.grid, I, unname
Loading required package: IRanges
Attaching package: 'IRanges'
The following object is masked from 'package:lubridate':
%within%
The following objects are masked from 'package:dplyr':
collapse, desc, slice
The following object is masked from 'package:purrr':
reduce
Loading required package: GenomeInfoDb
Loading required package: Biobase
Welcome to Bioconductor
Vignettes contain introductory material; view with 'browseVignettes()'. To cite Bioconductor, see 'citation("Biobase")', and for packages 'citation("pkgname")'.
Attaching package: 'Biobase'
The following object is masked from 'package:MatrixGenerics':
rowMedians
The following objects are masked from 'package:matrixStats':
anyMissing, rowMedianssce <- TENxPBMCData::TENxPBMCData('pbmc4k')see ?TENxPBMCData and browseVignettes('TENxPBMCData') for documentation
loading from cacherownames(sce) <- scuttle::uniquifyFeatureNames(rowData(sce)$ENSEMBL_ID, rowData(sce)$Symbol_TENx)
sceclass: SingleCellExperiment
dim: 33694 4340
metadata(0):
assays(1): counts
rownames(33694): RP11-34P13.3 FAM138A ... AC213203.1 FAM231B
rowData names(3): ENSEMBL_ID Symbol_TENx Symbol
colnames: NULL
colData names(11): Sample Barcode ... Individual Date_published
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):rowData(sce)DataFrame with 33694 rows and 3 columns
ENSEMBL_ID Symbol_TENx Symbol
<character> <character> <character>
RP11-34P13.3 ENSG00000243485 RP11-34P13.3 NA
FAM138A ENSG00000237613 FAM138A FAM138A
OR4F5 ENSG00000186092 OR4F5 OR4F5
RP11-34P13.7 ENSG00000238009 RP11-34P13.7 LOC100996442
RP11-34P13.8 ENSG00000239945 RP11-34P13.8 NA
... ... ... ...
AC233755.2 ENSG00000277856 AC233755.2 NA
AC233755.1 ENSG00000275063 AC233755.1 LOC102723407
AC240274.1 ENSG00000271254 AC240274.1 LOC102724250
AC213203.1 ENSG00000277475 AC213203.1 NA
FAM231B ENSG00000268674 FAM231B FAM231CcolData(sce)DataFrame with 4340 rows and 11 columns
Sample Barcode Sequence Library Cell_ranger_version Tissue_status Barcode_type Chemistry Sequence_platform Individual Date_published
<character> <character> <character> <integer> <character> <character> <character> <character> <character> <character> <character>
1 pbmc4k AAACCTGAGAAGGCCT-1 AAACCTGAGAAGGCCT 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
2 pbmc4k AAACCTGAGACAGACC-1 AAACCTGAGACAGACC 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
3 pbmc4k AAACCTGAGATAGTCA-1 AAACCTGAGATAGTCA 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
4 pbmc4k AAACCTGAGCGCCTCA-1 AAACCTGAGCGCCTCA 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
5 pbmc4k AAACCTGAGGCATGGT-1 AAACCTGAGGCATGGT 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
... ... ... ... ... ... ... ... ... ... ... ...
4336 pbmc4k TTTGGTTTCGCTAGCG-1 TTTGGTTTCGCTAGCG 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
4337 pbmc4k TTTGTCACACTTAACG-1 TTTGTCACACTTAACG 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
4338 pbmc4k TTTGTCACAGGTCCAC-1 TTTGTCACAGGTCCAC 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
4339 pbmc4k TTTGTCAGTTAAGACA-1 TTTGTCAGTTAAGACA 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08
4340 pbmc4k TTTGTCATCCCAAGAT-1 TTTGTCATCCCAAGAT 1 v2.1 NA Chromium Chromium_v2 Hiseq4000 HealthyDonor1 2017-11-08table(sce$Library)
1
4340