R
install.packages('mgcv')The estimated time for this lab is around 1h20.
R packages from different sources.R and Bioconductor.R and Bioconductor.Bioconductor and Seurat approaches to scRNAseq data analysis.R
“Hey, I’ve heard so many good things about this piece of software, it’s called ‘slingshot’? Heard of it? I really want to try it out on my dataset!!”
Or, in other words: “how do I install this or that brand new cutting-edge fancy package?”
R works with packages, available from different sources:
CRAN, the R developer team and official package provider: CRAN (which can probably win the title of “the worst webpage ever designed that survived until 2023”).Bioconductor, another package provider, with a primary focus on genomic-related packages: Bioconductor.GitHub.Let’s start by going over package installation.
Install mgcv, HCAData and revelio packages. Each of these three packages is available from a different source:
mgcv is a CRAN packageHCAData is a Bioconductor packagerevelio is a GitHub packageR
install.packages('mgcv')Installing package into ‘/home/rsg/R/x86_64-pc-linux-gnu-library/4.4’
(as ‘lib’ is unspecified)
trying URL 'https://cloud.r-project.org/src/contrib/mgcv_1.9-1.tar.gz'
Content type 'application/x-gzip' length 1083217 bytes (1.0 MB)
==================================================
downloaded 1.0 MB
* installing *source* package ‘mgcv’ ...
** package ‘mgcv’ successfully unpacked and MD5 sums checked
** using staged installation
** libs
using C compiler: ‘gcc (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0’
gcc -I"/usr/share/R/include" -DNDEBUG -fopenmp -fpic -g -O2 -fdebug-prefix-map=/build/r-base-4dpK2T/r-base-4.4.1=. -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -c coxph.c -o coxph.o
...
installing to /home/rsg/R/x86_64-pc-linux-gnu-library/4.4/00LOCK-mgcv/00new/mgcv/libs
** R
** data
** inst
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
** building package indices
** testing if installed package can be loaded from temporary location
** checking absolute paths in shared objects and dynamic libraries
** testing if installed package can be loaded from final location
** testing if installed package keeps a record of temporary installation path
* DONE (mgcv)
The downloaded source packages are in
‘/tmp/RtmpHmbVYD/downloaded_packages’R
BiocManager::install('HCAData')'getOption("repos")' replaces Bioconductor standard repositories, see 'help("repositories", package = "BiocManager")' for details.
Replacement repositories:
CRAN: https://cloud.r-project.org
Bioconductor version 3.22 (BiocManager 1.30.26), R 4.5.1 (2025-06-13)
Installing package(s) 'HCAData'
installing the source package ‘HCAData’
trying URL 'https://bioconductor.org/packages/3.22/data/experiment/src/contrib/HCAData_1.26.0.tar.gz'
Content type 'application/x-gzip' length 1547692 bytes (1.5 MB)
==================================================
downloaded 1.5 MB
R version 4.5.1 (2025-06-13) -- Great Square Root
* installing *source* package ‘HCAData’ ...
** this is package ‘HCAData’ version ‘1.26.0’
** using staged installation
** R
** inst
** byte-compile and prepare package for lazy loading
R version 4.5.1 (2025-06-13) -- Great Square Root
** help
*** installing help indices
** building package indices
R version 4.5.1 (2025-06-13) -- Great Square Root
** installing vignettes
** testing if installed package can be loaded from temporary location
R version 4.5.1 (2025-06-13) -- Great Square Root
** testing if installed package can be loaded from final location
R version 4.5.1 (2025-06-13) -- Great Square Root
** testing if installed package keeps a record of temporary installation path
* DONE (HCAData)
The downloaded source packages are in
‘/tmp/RtmpHmbVYD/downloaded_packages’R
devtools::install_github('danielschw188/Revelio')Using github PAT from envvar GITHUB_PAT. Use `gitcreds::gitcreds_set()` and unset GITHUB_PAT in .Renviron (or elsewhere) if you want to use the more secure git credential store instead.
Downloading GitHub repo danielschw188/Revelio@HEAD
These packages have more recent versions available.
It is recommended to update all of them.
...
** data
*** moving datasets to lazyload DB
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
** building package indices
** testing if installed package can be loaded from temporary location
** testing if installed package can be loaded from final location
** testing if installed package keeps a record of temporary installation path
* DONE (Revelio)Package help pages are available at different places, depending on their source. That being said, there is a place I like to go to easily find information related to most packages:
For instance, check out Revelio package help pages.
R and Bioconductor classesWhile CRAN is a repository of general-purpose packages, Bioconductor is the greatest source of analytical tools, data and workflows dedicated to genomic projects in R. Read more about Bioconductor to fully understand how it builds up on top of R general features, especially with the specific classes it introduces.
The two main concepts behind Bioconductor’s success are the non-redundant classes of objects it provides and their inter-operability. Huber et al., Nat. Methods 2015 summarizes it well.
tibble tables:tibbles are built on the fundamental data.frame objects. They follow “tidy” concepts, all gathered in a common tidyverse. This set of key concepts help general data investigation and data visualization through a set of associated packages such as ggplot2.
── Attaching core tidyverse packages ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.1 ✔ stringr 1.6.0
✔ ggplot2 4.0.1 ✔ tibble 3.3.0
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.2.0
── Conflicts ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors# A tibble: 5 × 4
x y z class
<int> <dbl> <dbl> <chr>
1 1 1 2 a
2 2 1 5 a
3 3 1 10 b
4 4 1 17 b
5 5 1 26 c tibblestibbles can be created from text files (or Excel files) using the readr package (part of tidyverse)
Rows: 32738 Columns: 2
── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: "\t"
chr (2): ID, Symbol
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.genes# A tibble: 32,738 × 2
ID Symbol
<chr> <chr>
1 ENSG00000243485 MIR1302-10
2 ENSG00000237613 FAM138A
3 ENSG00000186092 OR4F5
4 ENSG00000238009 RP11-34P13.7
5 ENSG00000239945 RP11-34P13.8
6 ENSG00000237683 AL627309.1
7 ENSG00000239906 RP11-34P13.14
8 ENSG00000241599 RP11-34P13.9
9 ENSG00000228463 AP006222.2
10 ENSG00000237094 RP4-669L17.10
# ℹ 32,728 more rowstibbles:tibbles can be readily “sliced” (i.e. selecting rows by number/name), “filtered” (i.e. selecting rows by condition) and columns can be “selected”. All these operations are performed using verbs (most of them provided by the dplyr package, part of tidyverse).
R
slice(genes, 1:4)# A tibble: 4 × 2
ID Symbol
<chr> <chr>
1 ENSG00000243485 MIR1302-10
2 ENSG00000237613 FAM138A
3 ENSG00000186092 OR4F5
4 ENSG00000238009 RP11-34P13.7filter(genes, Symbol == 'CCDC67')# A tibble: 1 × 2
ID Symbol
<chr> <chr>
1 ENSG00000165325 CCDC67# A tibble: 159 × 2
ID Symbol
<chr> <chr>
1 ENSG00000162592 CCDC27
2 ENSG00000160050 CCDC28B
3 ENSG00000186409 CCDC30
4 ENSG00000177868 CCDC23
5 ENSG00000159214 CCDC24
6 ENSG00000236624 CCDC163P
7 ENSG00000159588 CCDC17
8 ENSG00000122483 CCDC18
9 ENSG00000213085 CCDC19
10 ENSG00000117477 CCDC181
# ℹ 149 more rows# A tibble: 9 × 2
ID Symbol
<chr> <chr>
1 ENSG00000136710 CCDC115
2 ENSG00000183323 CCDC125
3 ENSG00000147419 CCDC25
4 ENSG00000149548 CCDC15
5 ENSG00000139537 CCDC65
6 ENSG00000151838 CCDC175
7 ENSG00000159625 CCDC135
8 ENSG00000160994 CCDC105
9 ENSG00000161609 CCDC155select(genes, 1)# A tibble: 32,738 × 1
ID
<chr>
1 ENSG00000243485
2 ENSG00000237613
3 ENSG00000186092
4 ENSG00000238009
5 ENSG00000239945
6 ENSG00000237683
7 ENSG00000239906
8 ENSG00000241599
9 ENSG00000228463
10 ENSG00000237094
# ℹ 32,728 more rowsselect(genes, ID)# A tibble: 32,738 × 1
ID
<chr>
1 ENSG00000243485
2 ENSG00000237613
3 ENSG00000186092
4 ENSG00000238009
5 ENSG00000239945
6 ENSG00000237683
7 ENSG00000239906
8 ENSG00000241599
9 ENSG00000228463
10 ENSG00000237094
# ℹ 32,728 more rows# A tibble: 32,738 × 1
Symbol
<chr>
1 MIR1302-10
2 FAM138A
3 OR4F5
4 RP11-34P13.7
5 RP11-34P13.8
6 AL627309.1
7 RP11-34P13.14
8 RP11-34P13.9
9 AP006222.2
10 RP4-669L17.10
# ℹ 32,728 more rowsColumns can also be quickly added/modified using the mutate verb.
# A tibble: 32,738 × 3
ID Symbol chr
<chr> <chr> <int>
1 ENSG00000243485 MIR1302-10 12
2 ENSG00000237613 FAM138A 9
3 ENSG00000186092 OR4F5 8
4 ENSG00000238009 RP11-34P13.7 17
5 ENSG00000239945 RP11-34P13.8 5
6 ENSG00000237683 AL627309.1 19
7 ENSG00000239906 RP11-34P13.14 17
8 ENSG00000241599 RP11-34P13.9 12
9 ENSG00000228463 AP006222.2 18
10 ENSG00000237094 RP4-669L17.10 2
# ℹ 32,728 more rows|> pipe:Actions on tibbles can be piped as a chain with |>, just like | pipes stdout as the stdin of the next command in bash. In this case, the first argument is always the output of the previous function and is ommited. Because tidyverse functions generally return a modified version of the input, pipping works remarkably well in such context.
R
Rows: 32738 Columns: 2
── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: "\t"
chr (2): ID, Symbol
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# A tibble: 3 × 1
ID
<chr>
1 ENSG00000152076
2 ENSG00000173421
3 ENSG00000024862SummarizedExperiment class:The most fundamental class used to hold the content of large-scale quantitative analyses, such as counts of RNA-seq experiments, or high-throughput cytometry experiments or proteomics experiments.
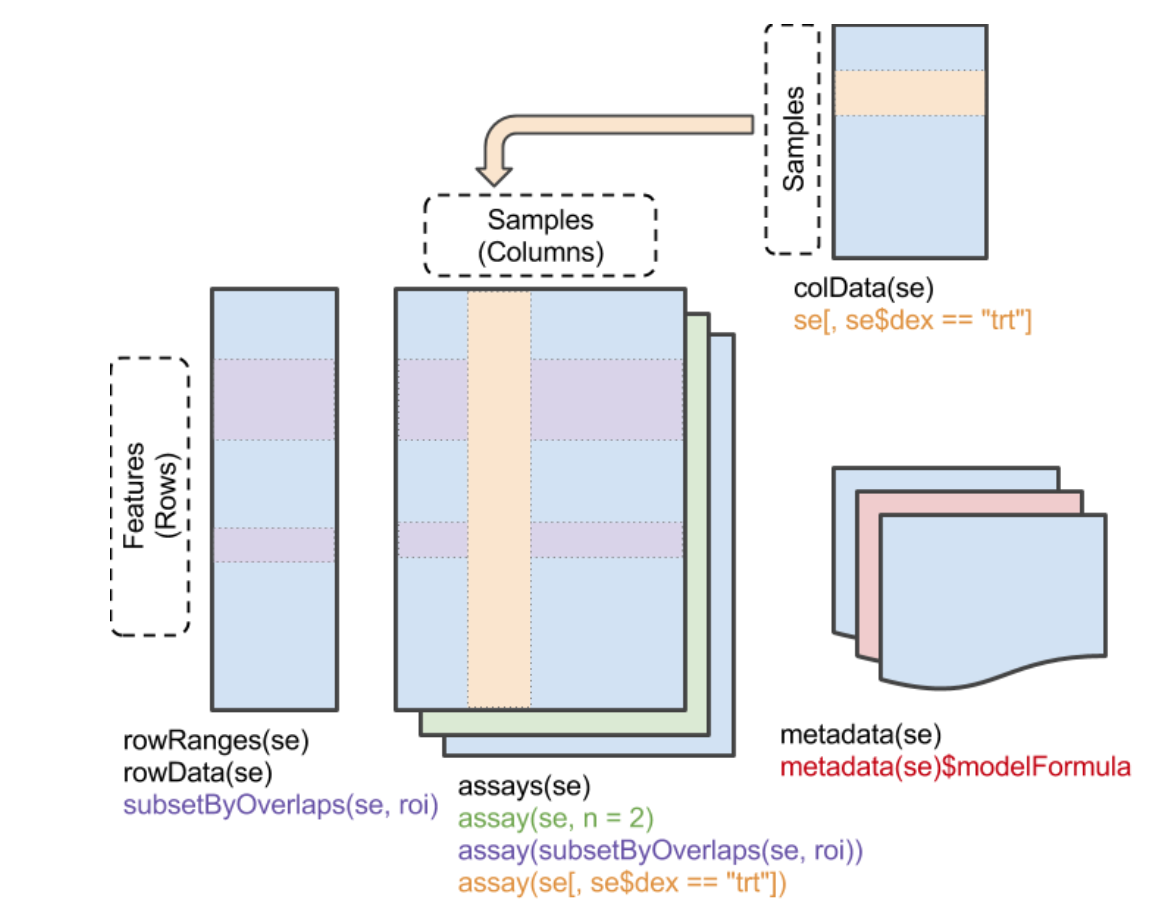
Make sure you understand the structure of objects from this class. A dedicated workshop that I would recommend quickly going over is available here. Generally speaking, a SummarizedExperiment object contains matrix-like objects (the assays), with rows representing features (e.g. genes, transcripts, …) and each column representing a sample. Information specific to genes and samples are stored in “parallel” data frames, for example to store gene locations, tissue of expression, biotypes (for genes) or batch, generation date, or machine ID (for samples). On top of that, metadata are also stored in the object (to store description of a project, …).
An important difference with S3 list-like objects usually used in R is that most of the underlying data (organized in precisely structured "slots") is accessed using getter functions, rather than the familiar $ or [. Here are some important getters:
assay(), assays(): Extrant matrix-like or list of matrix-like objects of identical dimensions. Since the objects are matrix-like, dim(), dimnames(), and 2-dimensional [, [<- methods are available.colData(): Annotations on each column (as a DataFrame): usually, description of each samplerowData(): Annotations on each row (as a DataFrame): usually, description of each genemetadata(): List of unstructured metadata describing the overall content of the object.Let’s dig into an example (you may need to install the airway package from Bioconductor…)
Loading required package: MatrixGenericsLoading required package: matrixStats
Attaching package: 'matrixStats'The following object is masked from 'package:dplyr':
count
Attaching package: 'MatrixGenerics'The following objects are masked from 'package:matrixStats':
colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse, colCounts, colCummaxs, colCummins, colCumprods, colCumsums, colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs, colMads, colMaxs, colMeans2,
colMedians, colMins, colOrderStats, colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds, colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads, colWeightedMeans, colWeightedMedians,
colWeightedSds, colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet, rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods, rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins, rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks, rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars, rowWeightedMads,
rowWeightedMeans, rowWeightedMedians, rowWeightedSds, rowWeightedVarsLoading required package: GenomicRangesLoading required package: stats4Loading required package: BiocGenericsLoading required package: generics
Attaching package: 'generics'The following object is masked from 'package:lubridate':
as.difftimeThe following object is masked from 'package:dplyr':
explainThe following objects are masked from 'package:base':
as.difftime, as.factor, as.ordered, intersect, is.element, setdiff, setequal, union
Attaching package: 'BiocGenerics'The following object is masked from 'package:dplyr':
combineThe following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
anyDuplicated, aperm, append, as.data.frame, basename, cbind, colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep, grepl, is.unsorted, lapply, Map, mapply, match, mget, order, paste,
pmax, pmax.int, pmin, pmin.int, Position, rank, rbind, Reduce, rownames, sapply, saveRDS, table, tapply, unique, unsplit, which.max, which.minLoading required package: S4Vectors
Attaching package: 'S4Vectors'The following objects are masked from 'package:lubridate':
second, second<-The following objects are masked from 'package:dplyr':
first, renameThe following object is masked from 'package:tidyr':
expandThe following object is masked from 'package:utils':
findMatchesThe following objects are masked from 'package:base':
expand.grid, I, unnameLoading required package: IRanges
Attaching package: 'IRanges'The following object is masked from 'package:lubridate':
%within%The following objects are masked from 'package:dplyr':
collapse, desc, sliceThe following object is masked from 'package:purrr':
reduceLoading required package: SeqinfoLoading required package: BiobaseWelcome to Bioconductor
Vignettes contain introductory material; view with 'browseVignettes()'. To cite Bioconductor, see 'citation("Biobase")', and for packages 'citation("pkgname")'.
Attaching package: 'Biobase'The following object is masked from 'package:MatrixGenerics':
rowMediansThe following objects are masked from 'package:matrixStats':
anyMissing, rowMediansclass: RangedSummarizedExperiment
dim: 63677 8
metadata(1): ''
assays(1): counts
rownames(63677): ENSG00000000003 ENSG00000000005 ... ENSG00000273492 ENSG00000273493
rowData names(10): gene_id gene_name ... seq_coord_system symbol
colnames(8): SRR1039508 SRR1039509 ... SRR1039520 SRR1039521
colData names(9): SampleName cell ... Sample BioSampleWhat are the dimensions of the dataset? What type of quantitative data is stored? Which features are assessed?
R
dim(airway)[1] 63677 8rowData(airway)DataFrame with 63677 rows and 10 columns
gene_id gene_name entrezid gene_biotype gene_seq_start gene_seq_end seq_name seq_strand seq_coord_system symbol
<character> <character> <integer> <character> <integer> <integer> <character> <integer> <integer> <character>
ENSG00000000003 ENSG00000000003 TSPAN6 NA protein_coding 99883667 99894988 X -1 NA TSPAN6
ENSG00000000005 ENSG00000000005 TNMD NA protein_coding 99839799 99854882 X 1 NA TNMD
ENSG00000000419 ENSG00000000419 DPM1 NA protein_coding 49551404 49575092 20 -1 NA DPM1
ENSG00000000457 ENSG00000000457 SCYL3 NA protein_coding 169818772 169863408 1 -1 NA SCYL3
ENSG00000000460 ENSG00000000460 C1orf112 NA protein_coding 169631245 169823221 1 1 NA C1orf112
... ... ... ... ... ... ... ... ... ... ...
ENSG00000273489 ENSG00000273489 RP11-180C16.1 NA antisense 131178723 131182453 7 -1 NA RP11-180C16.1
ENSG00000273490 ENSG00000273490 TSEN34 NA protein_coding 54693789 54697585 HSCHR19LRC_LRC_J_CTG1 1 NA TSEN34
ENSG00000273491 ENSG00000273491 RP11-138A9.2 NA lincRNA 130600118 130603315 HG1308_PATCH 1 NA RP11-138A9.2
ENSG00000273492 ENSG00000273492 AP000230.1 NA lincRNA 27543189 27589700 21 1 NA AP000230.1
ENSG00000273493 ENSG00000273493 RP11-80H18.4 NA lincRNA 58315692 58315845 3 1 NA RP11-80H18.4colData(airway)DataFrame with 8 rows and 9 columns
SampleName cell dex albut Run avgLength Experiment Sample BioSample
<factor> <factor> <factor> <factor> <factor> <integer> <factor> <factor> <factor>
SRR1039508 GSM1275862 N61311 untrt untrt SRR1039508 126 SRX384345 SRS508568 SAMN02422669
SRR1039509 GSM1275863 N61311 trt untrt SRR1039509 126 SRX384346 SRS508567 SAMN02422675
SRR1039512 GSM1275866 N052611 untrt untrt SRR1039512 126 SRX384349 SRS508571 SAMN02422678
SRR1039513 GSM1275867 N052611 trt untrt SRR1039513 87 SRX384350 SRS508572 SAMN02422670
SRR1039516 GSM1275870 N080611 untrt untrt SRR1039516 120 SRX384353 SRS508575 SAMN02422682
SRR1039517 GSM1275871 N080611 trt untrt SRR1039517 126 SRX384354 SRS508576 SAMN02422673
SRR1039520 GSM1275874 N061011 untrt untrt SRR1039520 101 SRX384357 SRS508579 SAMN02422683
SRR1039521 GSM1275875 N061011 trt untrt SRR1039521 98 SRX384358 SRS508580 SAMN02422677Can you create a subset of the data corresponding to LRG genes in untreated samples?
class: RangedSummarizedExperiment
dim: 0 4
metadata(1): ''
assays(1): counts
rownames(0):
rowData names(10): gene_id gene_name ... seq_coord_system symbol
colnames(4): SRR1039508 SRR1039512 SRR1039516 SRR1039520
colData names(9): SampleName cell ... Sample BioSampleGenomicRanges class (a.k.a. GRanges):GenomicRanges are a type of IntervalRanges, they are useful to describe genomic intervals. Each entry in a GRanges object has a seqnames(), a start() and an end() coordinates, a strand(), as well as associated metadata (mcols()). They can be built from scratch using tibbles converted with makeGRangesFromDataFrame().
R
library(GenomicRanges)
gr <- read_tsv('~/Share/GSM4486714_AXH009_genes.tsv', col_names = c('ID', 'Symbol')) |>
mutate(
chr = sample(1:22, n(), replace = TRUE),
start = sample(1:1000, n(), replace = TRUE),
end = sample(10000:20000, n(), replace = TRUE),
strand = sample(c('-', '+'), n(), replace = TRUE)
) |>
makeGRangesFromDataFrame(keep.extra.columns = TRUE)Rows: 32738 Columns: 2
── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: "\t"
chr (2): ID, Symbol
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.grGRanges object with 32738 ranges and 2 metadata columns:
seqnames ranges strand | ID Symbol
<Rle> <IRanges> <Rle> | <character> <character>
[1] 19 558-17646 + | ENSG00000243485 MIR1302-10
[2] 14 212-18716 + | ENSG00000237613 FAM138A
[3] 11 895-16915 - | ENSG00000186092 OR4F5
[4] 20 488-19429 - | ENSG00000238009 RP11-34P13.7
[5] 11 791-10309 + | ENSG00000239945 RP11-34P13.8
... ... ... ... . ... ...
[32734] 21 156-15929 - | ENSG00000215635 AC145205.1
[32735] 15 419-18539 + | ENSG00000268590 BAGE5
[32736] 15 607-19358 - | ENSG00000251180 CU459201.1
[32737] 8 176-11354 + | ENSG00000215616 AC002321.2
[32738] 10 787-16208 + | ENSG00000215611 AC002321.1
-------
seqinfo: 22 sequences from an unspecified genome; no seqlengthsmcols(gr)DataFrame with 32738 rows and 2 columns
ID Symbol
<character> <character>
1 ENSG00000243485 MIR1302-10
2 ENSG00000237613 FAM138A
3 ENSG00000186092 OR4F5
4 ENSG00000238009 RP11-34P13.7
5 ENSG00000239945 RP11-34P13.8
... ... ...
32734 ENSG00000215635 AC145205.1
32735 ENSG00000268590 BAGE5
32736 ENSG00000251180 CU459201.1
32737 ENSG00000215616 AC002321.2
32738 ENSG00000215611 AC002321.1Just like tidyverse in R, tidy functions are provided for GRanges by the plyranges package.
R
library(plyranges)
Attaching package: 'plyranges'The following object is masked from 'package:IRanges':
sliceThe following objects are masked from 'package:dplyr':
between, n, n_distinctThe following object is masked from 'package:stats':
filter
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22
195 194 170 159 184 190 170 186 176 166 166 161 190 202 175 184 173 194 164 151 161 183 Can you find a way to easily read common input files such as bed files into GRanges?
R
library(rtracklayer)
genes2 <- import('~/Share/GRCm39_genes.bed')
genes2GRanges object with 53700 ranges and 2 metadata columns:
seqnames ranges strand | name score
<Rle> <IRanges> <Rle> | <character> <numeric>
[1] 1 3143476-3144545 + | ENSMUSG00000102693 0
[2] 1 3172239-3172348 + | ENSMUSG00000064842 0
[3] 1 3276124-3741721 - | ENSMUSG00000051951 0
[4] 1 3322980-3323459 + | ENSMUSG00000102851 0
[5] 1 3435954-3438772 - | ENSMUSG00000103377 0
... ... ... ... . ... ...
[53696] Y 90763696-90766736 - | ENSMUSG00000095366 0
[53697] Y 90764326-90774754 + | ENSMUSG00000095134 0
[53698] Y 90796007-90827734 + | ENSMUSG00000096768 0
[53699] Y 90848682-90855309 + | ENSMUSG00000099871 0
[53700] Y 90850138-90850446 - | ENSMUSG00000096850 0
-------
seqinfo: 36 sequences from an unspecified genome; no seqlengthsHow would you have proceeded without rtracklayer? Check the start coordinates: what do you see? Comment on the interest of using Bioconductor.
R
Rows: 53700 Columns: 6
── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: "\t"
chr (3): X1, X4, X6
dbl (3): X2, X3, X5
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.genes2_manualGRanges object with 53700 ranges and 2 metadata columns:
seqnames ranges strand | id score
<Rle> <IRanges> <Rle> | <character> <numeric>
[1] 1 3143475-3144545 + | ENSMUSG00000102693 0
[2] 1 3172238-3172348 + | ENSMUSG00000064842 0
[3] 1 3276123-3741721 - | ENSMUSG00000051951 0
[4] 1 3322979-3323459 + | ENSMUSG00000102851 0
[5] 1 3435953-3438772 - | ENSMUSG00000103377 0
... ... ... ... . ... ...
[53696] Y 90763695-90766736 - | ENSMUSG00000095366 0
[53697] Y 90764325-90774754 + | ENSMUSG00000095134 0
[53698] Y 90796006-90827734 + | ENSMUSG00000096768 0
[53699] Y 90848681-90855309 + | ENSMUSG00000099871 0
[53700] Y 90850137-90850446 - | ENSMUSG00000096850 0
-------
seqinfo: 36 sequences from an unspecified genome; no seqlengths[1] 3143476 3172239 3276124 3322980 3435954 3445779[1] 3143475 3172238 3276123 3322979 3435953 3445778For single-cell RNA-seq projects, Bioconductor has been introducting new classes and standards very rapidly in the past few years. Notably, several packages are increasingly becoming central for single-cell analysis:
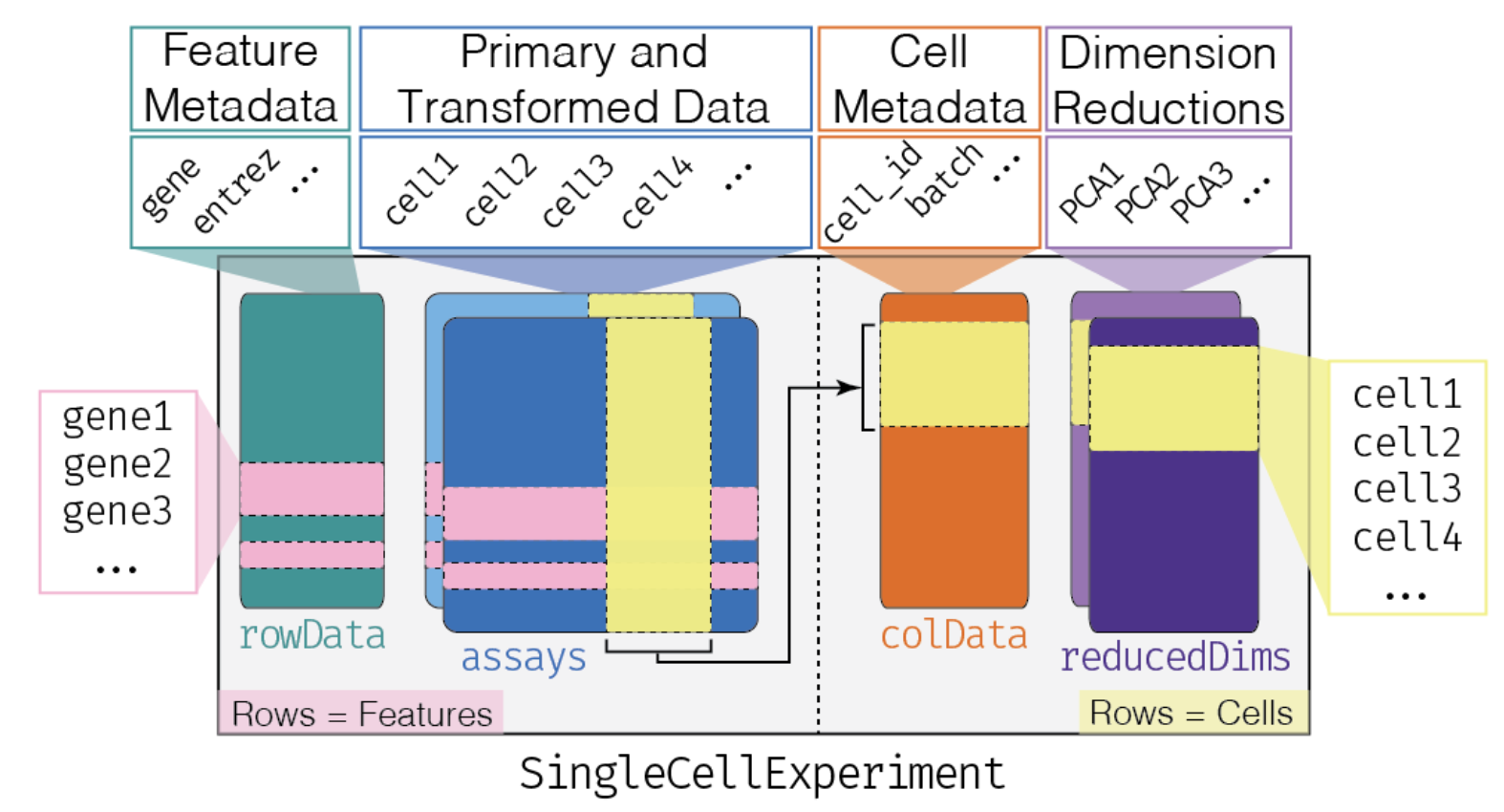
SingleCellExperimentscaterscranscuttlebatchelorSingleRblusterDropletUtilsslingshottradeSeqSingleCellExperiment is the fundamental class designed to contain single-cell (RNA-seq) data in Bioconductor ecosystem. It is a modified version of the SummarizedExperiment object, so most of the getters/setters are shared with this class.
R
loading from cachesceclass: SingleCellExperiment
dim: 500 1920
metadata(0):
assays(2): counts logcounts
rownames(500): ENSMUSG00000076609 ENSMUSG00000021250 ... ENSMUSG00000009566 ENSMUSG00000033809
rowData names(0):
colnames(1920): HSPC_007 HSPC_013 ... Prog_852 Prog_810
colData names(11): gate broad ... label sizeFactor
reducedDimNames(1): diffusion
mainExpName: endogenous
altExpNames(0):class(sce)[1] "SingleCellExperiment"
attr(,"package")
[1] "SingleCellExperiment"Several slots can be accessed in a SingleCellExperiment object, just like the SummarizedExperiment object it’s been adapted from:
R
colData(sce)DataFrame with 1920 rows and 11 columns
gate broad broad.mpp fine fine.mpp ESLAM HSC1 projected metrics label sizeFactor
<character> <character> <character> <character> <character> <logical> <logical> <logical> <DataFrame> <character> <numeric>
HSPC_007 HSPC NA NA NA NA FALSE FALSE FALSE 194829: 5022:0:... HSPC 0.0272469
HSPC_013 HSPC LMPP NA LMPP NA FALSE FALSE FALSE 110530: 15271:0:... HSPC 0.0904215
HSPC_019 HSPC LMPP NA NA NA FALSE FALSE FALSE 86825: 2708:0:... HSPC 0.0199211
HSPC_025 HSPC MPP MPP1 NA NA FALSE FALSE FALSE 212206:107278:0:... HSPC 0.5217920
HSPC_031 HSPC MPP STHSC NA NA FALSE FALSE FALSE 690411:227480:0:... HSPC 1.1062306
... ... ... ... ... ... ... ... ... ... ... ...
Prog_834 Prog CMP NA NA NA FALSE FALSE FALSE 471273:296060:0:... Prog 1.668383
Prog_840 Prog GMP NA GMP NA FALSE FALSE FALSE 421195:317394:0:... Prog 1.728596
Prog_846 Prog GMP NA NA NA FALSE FALSE FALSE 337564:253387:0:... Prog 1.282827
Prog_852 Prog MEP NA MEP NA FALSE FALSE FALSE 200193:136478:0:... Prog 0.643172
Prog_810 Prog CMP NA CMP NA FALSE FALSE FALSE 857257:396247:0:... Prog 2.614351rowData(sce)DataFrame with 500 rows and 0 columnsmetadata(sce)dim(sce)[1] 500 1920assays(sce)List of length 2
names(2): counts logcountsQuantitative metrics for scRNAseq studies can also be stored in assays:
R
assays(sce)List of length 2
names(2): counts logcountsassay(sce, 'counts')[1:10, 1:10]10 x 10 sparse Matrix of class "dgCMatrix" [[ suppressing 10 column names 'HSPC_007', 'HSPC_013', 'HSPC_019' ... ]]
ENSMUSG00000076609 30 16 7 17 19 11 1359 13 15 6
ENSMUSG00000021250 3 2 4 . 1 3 118 2 69 41
ENSMUSG00000076617 40 54 13 29 298 417 1107 9 16 15
ENSMUSG00000075602 3 248 7 537 640 7 300 1530 324 971
ENSMUSG00000006389 3 4 1 1171 6 2 271 5497 293 192
ENSMUSG00000041481 3 . 1 3 1 3 177 5 1 192
ENSMUSG00000024190 1 29 1 1733 1 5 20 3 1 1
ENSMUSG00000003949 4 15 3 175 915 1131 41 1465 330 258
ENSMUSG00000052684 1 4 2 144 82 5 142 578 . 78
ENSMUSG00000026358 . 27 6 344 3 9 3 3612 185 .assay(sce, 'logcounts')[1:10, 1:10]10 x 10 sparse Matrix of class "dgCMatrix" [[ suppressing 10 column names 'HSPC_007', 'HSPC_013', 'HSPC_019' ... ]]
ENSMUSG00000076609 10.105963 7.475320 8.461015 5.069532 4.1839186 4.484632 13.057990 2.7927556 4.907536 4.256554
ENSMUSG00000021250 6.795769 4.530984 7.656727 . 0.9290112 2.772518 9.534081 0.9352591 7.071052 6.963227
ENSMUSG00000076617 10.520673 9.224491 9.352209 5.822162 8.0788620 9.664957 12.762138 2.3519259 4.997638 5.532469
ENSMUSG00000075602 6.795769 11.421912 8.461015 10.008633 9.1787674 3.868916 10.879076 9.4488576 9.293908 11.517886
ENSMUSG00000006389 6.795769 5.499439 5.678018 11.132621 2.6834323 2.289440 10.732488 11.2924799 9.149058 9.181516
ENSMUSG00000041481 6.795769 . 5.678018 2.754763 0.9290112 2.772518 10.118395 1.7139531 1.552981 9.181516
ENSMUSG00000024190 5.236545 8.329662 5.678018 11.697944 0.9290112 3.422457 6.982870 1.2438829 1.552981 2.006833
ENSMUSG00000003949 7.207556 7.382751 7.244071 8.393960 9.6937189 11.103314 8.012640 9.3863182 9.320338 9.607145
ENSMUSG00000052684 5.236545 5.499439 6.663859 8.113597 6.2312326 3.422457 9.800857 8.0478635 . 7.885582
ENSMUSG00000026358 . 8.226901 8.239305 9.366905 1.8921625 4.209375 4.309131 10.6869334 8.487165 . counts(sce)[1:10, 1:10]10 x 10 sparse Matrix of class "dgCMatrix" [[ suppressing 10 column names 'HSPC_007', 'HSPC_013', 'HSPC_019' ... ]]
ENSMUSG00000076609 30 16 7 17 19 11 1359 13 15 6
ENSMUSG00000021250 3 2 4 . 1 3 118 2 69 41
ENSMUSG00000076617 40 54 13 29 298 417 1107 9 16 15
ENSMUSG00000075602 3 248 7 537 640 7 300 1530 324 971
ENSMUSG00000006389 3 4 1 1171 6 2 271 5497 293 192
ENSMUSG00000041481 3 . 1 3 1 3 177 5 1 192
ENSMUSG00000024190 1 29 1 1733 1 5 20 3 1 1
ENSMUSG00000003949 4 15 3 175 915 1131 41 1465 330 258
ENSMUSG00000052684 1 4 2 144 82 5 142 578 . 78
ENSMUSG00000026358 . 27 6 344 3 9 3 3612 185 .logcounts(sce)[1:10, 1:10]10 x 10 sparse Matrix of class "dgCMatrix" [[ suppressing 10 column names 'HSPC_007', 'HSPC_013', 'HSPC_019' ... ]]
ENSMUSG00000076609 10.105963 7.475320 8.461015 5.069532 4.1839186 4.484632 13.057990 2.7927556 4.907536 4.256554
ENSMUSG00000021250 6.795769 4.530984 7.656727 . 0.9290112 2.772518 9.534081 0.9352591 7.071052 6.963227
ENSMUSG00000076617 10.520673 9.224491 9.352209 5.822162 8.0788620 9.664957 12.762138 2.3519259 4.997638 5.532469
ENSMUSG00000075602 6.795769 11.421912 8.461015 10.008633 9.1787674 3.868916 10.879076 9.4488576 9.293908 11.517886
ENSMUSG00000006389 6.795769 5.499439 5.678018 11.132621 2.6834323 2.289440 10.732488 11.2924799 9.149058 9.181516
ENSMUSG00000041481 6.795769 . 5.678018 2.754763 0.9290112 2.772518 10.118395 1.7139531 1.552981 9.181516
ENSMUSG00000024190 5.236545 8.329662 5.678018 11.697944 0.9290112 3.422457 6.982870 1.2438829 1.552981 2.006833
ENSMUSG00000003949 7.207556 7.382751 7.244071 8.393960 9.6937189 11.103314 8.012640 9.3863182 9.320338 9.607145
ENSMUSG00000052684 5.236545 5.499439 6.663859 8.113597 6.2312326 3.422457 9.800857 8.0478635 . 7.885582
ENSMUSG00000026358 . 8.226901 8.239305 9.366905 1.8921625 4.209375 4.309131 10.6869334 8.487165 . Check the colData() output of the sce object. What information is stored there? How can you access the different objects stored in colData?
R
colData(sce)DataFrame with 1920 rows and 11 columns
gate broad broad.mpp fine fine.mpp ESLAM HSC1 projected metrics label sizeFactor
<character> <character> <character> <character> <character> <logical> <logical> <logical> <DataFrame> <character> <numeric>
HSPC_007 HSPC NA NA NA NA FALSE FALSE FALSE 194829: 5022:0:... HSPC 0.0272469
HSPC_013 HSPC LMPP NA LMPP NA FALSE FALSE FALSE 110530: 15271:0:... HSPC 0.0904215
HSPC_019 HSPC LMPP NA NA NA FALSE FALSE FALSE 86825: 2708:0:... HSPC 0.0199211
HSPC_025 HSPC MPP MPP1 NA NA FALSE FALSE FALSE 212206:107278:0:... HSPC 0.5217920
HSPC_031 HSPC MPP STHSC NA NA FALSE FALSE FALSE 690411:227480:0:... HSPC 1.1062306
... ... ... ... ... ... ... ... ... ... ... ...
Prog_834 Prog CMP NA NA NA FALSE FALSE FALSE 471273:296060:0:... Prog 1.668383
Prog_840 Prog GMP NA GMP NA FALSE FALSE FALSE 421195:317394:0:... Prog 1.728596
Prog_846 Prog GMP NA NA NA FALSE FALSE FALSE 337564:253387:0:... Prog 1.282827
Prog_852 Prog MEP NA MEP NA FALSE FALSE FALSE 200193:136478:0:... Prog 0.643172
Prog_810 Prog CMP NA CMP NA FALSE FALSE FALSE 857257:396247:0:... Prog 2.614351$gate
[1] "character"
$broad
[1] "character"
$broad.mpp
[1] "character"
$fine
[1] "character"
$fine.mpp
[1] "character"
$ESLAM
[1] "logical"
$HSC1
[1] "logical"
$projected
[1] "logical"
$metrics
[1] "DFrame"
attr(,"package")
[1] "S4Vectors"
$label
[1] "character"
$sizeFactor
[1] "numeric"[1] "HSPC" "HSPC" "HSPC" "HSPC" "HSPC" "HSPC"NULLhead(sce$sizeFactor) HSPC_007 HSPC_013 HSPC_019 HSPC_025 HSPC_031 HSPC_037
0.02724693 0.09042153 0.01992108 0.52179199 1.10623062 0.51431538 Are there any reduced dimensionality representation of the data stored in the sce object? How can we run a PCA using normalized counts?
R
reducedDims(sce)List of length 1
names(1): diffusion[1] "sdev" "rotation" "center" "scale" "x" dim(pca$x)[1] 1920 500head(pca$x[, 1:50]) PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PC10 PC11 PC12 PC13 PC14 PC15 PC16 PC17 PC18 PC19 PC20 PC21
HSPC_007 5.581553 -3.4100947 9.951168 13.0797762 3.545895 10.1226017 -4.1159055 -3.4238633 -7.936727 10.6921786 -1.048376 -4.362441 5.252001 1.7514129 -0.9191112 -4.720843 -2.9529841 1.72653427 -1.1546955 5.967443 -4.9219043
HSPC_013 -12.193370 -5.3973727 1.573549 13.7750818 -1.316799 -0.9443251 -0.4590312 1.4407575 -4.399518 8.6381255 -3.892309 -3.310407 2.193760 6.3337138 0.6321951 1.247678 2.5258067 -2.27899711 8.3330827 1.203497 0.5654264
HSPC_019 -3.528223 7.7885911 5.305870 -0.6928286 4.046022 0.3507853 -5.8048713 -5.5745236 -4.817877 6.9710785 -4.621007 -2.086735 10.624674 0.1293415 -6.3774706 7.536987 1.3668570 -0.07492649 -0.9040375 -1.573461 -1.3513760
HSPC_025 -25.082329 5.2035295 4.627378 14.0209886 -1.869520 -5.9116901 -0.1576345 5.7150177 2.998934 4.4459935 0.259513 2.635814 -6.254529 3.3289181 1.9856744 -5.554629 10.6416059 -9.62351660 -2.4473404 1.696153 -5.6186977
HSPC_031 -16.159834 8.0498872 -10.116348 4.2919683 2.683031 -6.7923050 8.0293846 -1.9657941 -2.445369 0.9570117 -5.458279 -12.344929 5.930111 -5.9945465 -0.7331365 2.682199 1.3677304 -9.98871025 4.5417044 -6.343339 1.8645220
HSPC_037 -20.450257 -0.6194833 -7.510092 14.1745410 8.051961 1.0421148 10.5405868 0.7380758 -3.666130 -0.3892389 -10.137915 1.092324 6.488297 2.9905900 3.6858905 -9.673307 0.5212146 -2.05971416 -1.9071441 7.463945 -10.3870858
PC22 PC23 PC24 PC25 PC26 PC27 PC28 PC29 PC30 PC31 PC32 PC33 PC34 PC35 PC36 PC37 PC38 PC39 PC40 PC41 PC42
HSPC_007 2.9618115 -5.5791148 -2.268495 1.095512 -2.3194333 -6.8060168 1.4734275 4.6384342 -1.3155950 -5.7868406 -1.925339 1.104915 -4.904943 -5.7231568 7.0623893 -5.456147 0.3217448 -2.572326 -4.551055 -0.1726143 5.842971
HSPC_013 4.9754802 -0.5438859 -2.799659 4.265413 0.4714923 -0.4501818 0.7892971 2.6084879 -5.2467188 0.5364803 1.722019 4.990867 4.896010 1.2722998 -3.8090271 -1.647040 -2.6976177 1.103188 -2.356270 1.6141577 -3.259530
HSPC_019 4.0940259 0.8512859 -1.442058 10.897733 6.1569133 1.4970447 4.0109183 -11.2219306 -6.4404311 -2.7100811 2.059306 -4.575883 -2.260518 -8.2634335 -2.3966379 2.999743 -3.4977372 -4.638447 1.270189 3.3668990 5.400202
HSPC_025 2.7414756 0.3282526 -1.698754 -7.288994 -0.8159283 2.8763124 -3.5496455 0.1712682 2.6777455 -5.5417113 -1.859348 -0.757561 -5.178513 -0.5872237 -0.8708151 2.772968 -5.2629035 -4.497201 -2.013552 -4.0707202 -1.072854
HSPC_031 -0.9663505 -5.8157882 6.189554 2.518689 9.3543754 6.1909333 2.5499150 4.2883360 -0.7154207 -8.2774092 -3.288322 -11.886294 -4.332450 6.2491786 3.0642466 3.002628 -11.6083495 -15.142854 -1.293257 -12.9505869 -11.287041
HSPC_037 1.4925686 -4.0960445 -3.744462 5.418505 -7.6114089 -3.4052990 -0.8200447 0.2419793 3.4306565 -0.3564786 -5.354014 -8.363324 3.751114 -7.3113139 0.2295738 4.045629 -6.4420490 1.771556 -3.322189 -6.1165457 -1.213184
PC43 PC44 PC45 PC46 PC47 PC48 PC49 PC50
HSPC_007 -3.50678908 3.4892276 5.848641 -0.2128977 0.4942121 -0.07747664 -2.827778 2.412009
HSPC_013 -5.03456958 1.1780332 -1.552541 -2.3808517 -2.9659444 -0.74742282 1.149531 2.472806
HSPC_019 -3.12221617 3.2583780 1.817843 -3.1729495 -1.1888832 -0.59825631 -9.962968 -2.585295
HSPC_025 -2.77454846 0.4562519 -6.480854 0.6699326 -5.8678726 0.17369337 7.514302 1.163721
HSPC_031 0.09341584 3.9700545 2.692311 -3.1757431 1.6065308 -1.97332056 -3.111541 -5.190516
HSPC_037 3.59481984 7.5068703 1.282379 -7.0477980 3.7969450 1.60118178 -5.204884 9.217837We will see more advanced ways of reducing scRNAseq data dimensionality in the coming lectures and labs.
Seurat is another very popular ecosystem to investigate scRNAseq data. It is primarily developed and maintained by the Sajita Lab. It originally begun as a single package aiming at encompassing “all” (most) aspects of scRNAseq analysis. However, it rapidly evolved in a much larger project, and now operates along with other “wrappers” and extensions. It also has a very extended support from the lab group. All in all, is provides a (somewhat) simple workflow to start investigating scRNAseq data.
It is important to chose one standard that you feel comfortable with yourself. Which standard provides the most intuitive approach for you? Do you prefer an “all-in-one, plug-n-play” workflow (Seurat-style), or a modular approach (Bioconductor-style)? Which documentation is easier to read for you, a central full-featured website with extensive examples (Seurat-style), or “programmatic”-style vignettes (Bioconductor-style)?
This course will mostly rely on Bioconductor-based methods, but sometimes use Seurat-based methods.s In the absence of coordination of data structures, the next best solution is to write functions to coerce an object from a certain class to another class (i.e. Seurat to SingleCellExperiment, or vice-versa). Luckily, this is quite straightforward in R for these 2 data classes:
R
sce_seurat <- Seurat::as.Seurat(sce)Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Warning: Keys should be one or more alphanumeric characters followed by an underscore, setting key from DC to DC_sceclass: SingleCellExperiment
dim: 500 1920
metadata(0):
assays(2): counts logcounts
rownames(500): ENSMUSG00000076609 ENSMUSG00000021250 ... ENSMUSG00000009566 ENSMUSG00000033809
rowData names(0):
colnames(1920): HSPC_007 HSPC_013 ... Prog_852 Prog_810
colData names(11): gate broad ... label sizeFactorFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'reducedDimNames(1): diffusion
mainExpName: endogenousFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'altExpNames(0):sce_seuratAn object of class Seurat
500 features across 1920 samples within 1 assay
Active assay: endogenous (500 features, 0 variable features)
2 layers present: counts, data
1 dimensional reduction calculated: diffusionsce2 <- Seurat::as.SingleCellExperiment(sce_seurat)Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Do you see any change between sce and the corresponding, “back-converted”, sce2 objects? Explain these differences.
R
sceclass: SingleCellExperiment
dim: 500 1920
metadata(0):
assays(2): counts logcounts
rownames(500): ENSMUSG00000076609 ENSMUSG00000021250 ... ENSMUSG00000009566 ENSMUSG00000033809
rowData names(0):
colnames(1920): HSPC_007 HSPC_013 ... Prog_852 Prog_810
colData names(11): gate broad ... label sizeFactorFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'reducedDimNames(1): diffusion
mainExpName: endogenousFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'altExpNames(0):sce2class: SingleCellExperiment
dim: 500 1920
metadata(0):
assays(2): counts logcounts
rownames(500): ENSMUSG00000076609 ENSMUSG00000021250 ... ENSMUSG00000009566 ENSMUSG00000033809
rowData names(0):
colnames(1920): HSPC_007 HSPC_013 ... Prog_852 Prog_810
colData names(19): orig.ident nCount_endogenous ... sizeFactor identFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'reducedDimNames(1): DIFFUSION
mainExpName: endogenousFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'altExpNames(0):#
colData(sce)DataFrame with 1920 rows and 11 columns
gate broad broad.mpp fine fine.mpp ESLAM HSC1 projected metrics label sizeFactor
<character> <character> <character> <character> <character> <logical> <logical> <logical> <DataFrame> <character> <numeric>
HSPC_007 HSPC NA NA NA NA FALSE FALSE FALSE 194829: 5022:0:... HSPC 0.0272469
HSPC_013 HSPC LMPP NA LMPP NA FALSE FALSE FALSE 110530: 15271:0:... HSPC 0.0904215
HSPC_019 HSPC LMPP NA NA NA FALSE FALSE FALSE 86825: 2708:0:... HSPC 0.0199211
HSPC_025 HSPC MPP MPP1 NA NA FALSE FALSE FALSE 212206:107278:0:... HSPC 0.5217920
HSPC_031 HSPC MPP STHSC NA NA FALSE FALSE FALSE 690411:227480:0:... HSPC 1.1062306
... ... ... ... ... ... ... ... ... ... ... ...
Prog_834 Prog CMP NA NA NA FALSE FALSE FALSE 471273:296060:0:... Prog 1.668383
Prog_840 Prog GMP NA GMP NA FALSE FALSE FALSE 421195:317394:0:... Prog 1.728596
Prog_846 Prog GMP NA NA NA FALSE FALSE FALSE 337564:253387:0:... Prog 1.282827
Prog_852 Prog MEP NA MEP NA FALSE FALSE FALSE 200193:136478:0:... Prog 0.643172
Prog_810 Prog CMP NA CMP NA FALSE FALSE FALSE 857257:396247:0:... Prog 2.614351colData(sce2)DataFrame with 1920 rows and 19 columns
orig.ident nCount_endogenous nFeature_endogenous gate broad broad.mpp fine fine.mpp ESLAM HSC1 projected metrics.X__no_feature metrics.X__ambiguous metrics.X__too_low_aQual metrics.X__not_aligned
<factor> <numeric> <integer> <character> <character> <character> <character> <character> <logical> <logical> <logical> <numeric> <numeric> <numeric> <numeric>
HSPC_007 HSPC 1262 381 HSPC NA NA NA NA FALSE FALSE FALSE 194829 5022 0 5820455
HSPC_013 HSPC 5906 409 HSPC LMPP NA LMPP NA FALSE FALSE FALSE 110530 15271 0 1562724
HSPC_019 HSPC 1002 355 HSPC LMPP NA NA NA FALSE FALSE FALSE 86825 2708 0 1407254
HSPC_025 HSPC 58177 434 HSPC MPP MPP1 NA NA FALSE FALSE FALSE 212206 107278 0 1810368
HSPC_031 HSPC 118570 435 HSPC MPP STHSC NA NA FALSE FALSE FALSE 690411 227480 0 6097116
... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ...
Prog_834 Prog 185771 476 Prog CMP NA NA NA FALSE FALSE FALSE 471273 296060 0 4312094
Prog_840 Prog 245841 484 Prog GMP NA GMP NA FALSE FALSE FALSE 421195 317394 0 3770447
Prog_846 Prog 199277 483 Prog GMP NA NA NA FALSE FALSE FALSE 337564 253387 0 2549458
Prog_852 Prog 56543 464 Prog MEP NA MEP NA FALSE FALSE FALSE 200193 136478 0 1442745
Prog_810 Prog 266651 480 Prog CMP NA CMP NA FALSE FALSE FALSE 857257 396247 0 4518455
metrics.X__alignment_not_unique label sizeFactor ident
<numeric> <character> <numeric> <factor>
HSPC_007 0 HSPC 0.0272469 SingleCellExperiment
HSPC_013 0 HSPC 0.0904215 SingleCellExperiment
HSPC_019 0 HSPC 0.0199211 SingleCellExperiment
HSPC_025 0 HSPC 0.5217920 SingleCellExperiment
HSPC_031 0 HSPC 1.1062306 SingleCellExperiment
... ... ... ... ...
Prog_834 0 Prog 1.668383 SingleCellExperiment
Prog_840 0 Prog 1.728596 SingleCellExperiment
Prog_846 0 Prog 1.282827 SingleCellExperiment
Prog_852 0 Prog 0.643172 SingleCellExperiment
Prog_810 0 Prog 2.614351 SingleCellExperimentTry and access the underlying raw or normalized data from the sce_seurat object. How does it compare to data access from an SingleCellExperiment object?
R
colnames(sce_seurat) [1] "HSPC_007" "HSPC_013" "HSPC_019" "HSPC_025" "HSPC_031" "HSPC_037" "LT-HSC_001" "HSPC_001" "HSPC_008" "HSPC_014" "HSPC_020" "HSPC_026" "HSPC_032" "HSPC_038" "LT-HSC_002" "HSPC_002" "HSPC_009" "HSPC_015"
[19] "HSPC_021" "HSPC_027" "HSPC_033" "HSPC_039" "LT-HSC_003" "HSPC_003" "HSPC_010" "HSPC_016" "HSPC_022" "HSPC_028" "HSPC_034" "HSPC_040" "LT-HSC_004" "HSPC_004" "HSPC_011" "HSPC_017" "HSPC_023" "HSPC_029"
[37] "HSPC_035" "HSPC_041" "LT-HSC_005" "HSPC_005" "HSPC_012" "HSPC_018" "HSPC_024" "HSPC_030" "HSPC_036" "HSPC_042" "LT-HSC_006" "HSPC_006" "Prog_007" "Prog_013" "Prog_019" "Prog_025" "Prog_031" "Prog_037"
[55] "LT-HSC_007" "Prog_001" "Prog_008" "Prog_014" "Prog_020" "Prog_026" "Prog_032" "Prog_038" "LT-HSC_008" "Prog_002" "Prog_009" "Prog_015" "Prog_021" "Prog_027" "Prog_033" "Prog_039" "LT-HSC_009" "Prog_003"
[73] "Prog_010" "Prog_016" "Prog_022" "Prog_028" "Prog_034" "Prog_040" "LT-HSC_010" "Prog_004" "Prog_011" "Prog_017" "Prog_023" "Prog_029" "Prog_035" "Prog_041" "LT-HSC_011" "Prog_005" "Prog_012" "Prog_018"
[91] "Prog_024" "Prog_030" "Prog_036" "Prog_042" "LT-HSC_012" "Prog_006" "HSPC_049" "HSPC_055" "HSPC_061" "HSPC_067" "HSPC_073" "HSPC_079" "LT-HSC_013" "HSPC_043" "HSPC_050" "HSPC_056" "HSPC_062" "HSPC_068"
[109] "HSPC_074" "HSPC_080" "LT-HSC_014" "HSPC_044" "HSPC_051" "HSPC_057" "HSPC_063" "HSPC_069" "HSPC_075" "HSPC_081" "LT-HSC_015" "HSPC_045" "HSPC_052" "HSPC_058" "HSPC_064" "HSPC_070" "HSPC_076" "HSPC_082"
[127] "LT-HSC_016" "HSPC_046" "HSPC_053" "HSPC_059" "HSPC_065" "HSPC_071" "HSPC_077" "HSPC_083" "LT-HSC_017" "HSPC_047" "HSPC_054" "HSPC_060" "HSPC_066" "HSPC_072" "HSPC_078" "HSPC_084" "LT-HSC_018" "HSPC_048"
[145] "Prog_049" "Prog_055" "Prog_061" "Prog_067" "Prog_073" "Prog_079" "LT-HSC_019" "Prog_043" "Prog_050" "Prog_056" "Prog_062" "Prog_068" "Prog_074" "Prog_080" "LT-HSC_020" "Prog_044" "Prog_051" "Prog_057"
[163] "Prog_063" "Prog_069" "Prog_075" "Prog_081" "LT-HSC_021" "Prog_045" "Prog_052" "Prog_058" "Prog_064" "Prog_070" "Prog_076" "Prog_082" "LT-HSC_022" "Prog_046" "Prog_053" "Prog_059" "Prog_065" "Prog_071"
[181] "Prog_077" "Prog_083" "LT-HSC_023" "Prog_047" "Prog_054" "Prog_060" "Prog_066" "Prog_072" "Prog_078" "Prog_084" "LT-HSC_024" "Prog_048" "HSPC_091" "HSPC_097" "HSPC_103" "HSPC_109" "HSPC_115" "HSPC_121"
[199] "LT-HSC_025" "HSPC_085" "HSPC_092" "HSPC_098" "HSPC_104" "HSPC_110" "HSPC_116" "HSPC_122" "LT-HSC_026" "HSPC_086" "HSPC_093" "HSPC_099" "HSPC_105" "HSPC_111" "HSPC_117" "HSPC_123" "LT-HSC_027" "HSPC_087"
[217] "HSPC_094" "HSPC_100" "HSPC_106" "HSPC_112" "HSPC_118" "HSPC_124" "LT-HSC_028" "HSPC_088" "HSPC_095" "HSPC_101" "HSPC_107" "HSPC_113" "HSPC_119" "HSPC_125" "LT-HSC_029" "HSPC_089" "HSPC_096" "HSPC_102"
[235] "HSPC_108" "HSPC_114" "HSPC_120" "HSPC_126" "LT-HSC_030" "HSPC_090" "Prog_091" "Prog_097" "Prog_103" "Prog_109" "Prog_115" "Prog_121" "LT-HSC_031" "Prog_085" "Prog_092" "Prog_098" "Prog_104" "Prog_110"
[253] "Prog_116" "Prog_122" "LT-HSC_032" "Prog_086" "Prog_093" "Prog_099" "Prog_105" "Prog_111" "Prog_117" "Prog_123" "LT-HSC_033" "Prog_087" "Prog_094" "Prog_100" "Prog_106" "Prog_112" "Prog_118" "Prog_124"
[271] "LT-HSC_034" "Prog_088" "Prog_095" "Prog_101" "Prog_107" "Prog_113" "Prog_119" "Prog_125" "LT-HSC_035" "Prog_089" "Prog_096" "Prog_102" "Prog_108" "Prog_114" "Prog_120" "Prog_126" "LT-HSC_036" "Prog_090"
[289] "HSPC_133" "HSPC_139" "HSPC_145" "HSPC_151" "HSPC_157" "HSPC_163" "LT-HSC_037" "HSPC_127" "HSPC_134" "HSPC_140" "HSPC_146" "HSPC_152" "HSPC_158" "HSPC_164" "LT-HSC_038" "HSPC_128" "HSPC_135" "HSPC_141"
[307] "HSPC_147" "HSPC_153" "HSPC_159" "HSPC_165" "LT-HSC_039" "HSPC_129" "HSPC_136" "HSPC_142" "HSPC_148" "HSPC_154" "HSPC_160" "HSPC_166" "LT-HSC_040" "HSPC_130" "HSPC_137" "HSPC_143" "HSPC_149" "HSPC_155"
[325] "HSPC_161" "HSPC_167" "LT-HSC_041" "HSPC_131" "HSPC_138" "HSPC_144" "HSPC_150" "HSPC_156" "HSPC_162" "HSPC_168" "LT-HSC_042" "HSPC_132" "Prog_133" "Prog_139" "Prog_145" "Prog_151" "Prog_157" "Prog_163"
[343] "LT-HSC_043" "Prog_127" "Prog_134" "Prog_140" "Prog_146" "Prog_152" "Prog_158" "Prog_164" "LT-HSC_044" "Prog_128" "Prog_135" "Prog_141" "Prog_147" "Prog_153" "Prog_159" "Prog_165" "LT-HSC_045" "Prog_129"
[361] "Prog_136" "Prog_142" "Prog_148" "Prog_154" "Prog_160" "Prog_166" "LT-HSC_046" "Prog_130" "Prog_137" "Prog_143" "Prog_149" "Prog_155" "Prog_161" "Prog_167" "LT-HSC_047" "Prog_131" "Prog_138" "Prog_144"
[379] "Prog_150" "Prog_156" "Prog_162" "Prog_168" "LT-HSC_048" "Prog_132" "HSPC_175" "HSPC_181" "HSPC_187" "HSPC_193" "HSPC_199" "HSPC_205" "LT-HSC_049" "HSPC_169" "HSPC_176" "HSPC_182" "HSPC_188" "HSPC_194"
[397] "HSPC_200" "HSPC_206" "LT-HSC_050" "HSPC_170" "HSPC_177" "HSPC_183" "HSPC_189" "HSPC_195" "HSPC_201" "HSPC_207" "LT-HSC_051" "HSPC_171" "HSPC_178" "HSPC_184" "HSPC_190" "HSPC_196" "HSPC_202" "HSPC_208"
[415] "LT-HSC_052" "HSPC_172" "HSPC_179" "HSPC_185" "HSPC_191" "HSPC_197" "HSPC_203" "HSPC_209" "LT-HSC_053" "HSPC_173" "HSPC_180" "HSPC_186" "HSPC_192" "HSPC_198" "HSPC_204" "HSPC_210" "LT-HSC_054" "HSPC_174"
[433] "Prog_175" "Prog_181" "Prog_187" "Prog_193" "Prog_199" "Prog_205" "LT-HSC_055" "Prog_169" "Prog_176" "Prog_182" "Prog_188" "Prog_194" "Prog_200" "Prog_206" "LT-HSC_056" "Prog_170" "Prog_177" "Prog_183"
[451] "Prog_189" "Prog_195" "Prog_201" "Prog_207" "LT-HSC_057" "Prog_171" "Prog_178" "Prog_184" "Prog_190" "Prog_196" "Prog_202" "Prog_208" "LT-HSC_058" "Prog_172" "Prog_179" "Prog_185" "Prog_191" "Prog_197"
[469] "Prog_203" "Prog_209" "LT-HSC_059" "Prog_173" "Prog_180" "Prog_186" "Prog_192" "Prog_198" "Prog_204" "Prog_210" "LT-HSC_060" "Prog_174" "HSPC_217" "HSPC_223" "HSPC_229" "HSPC_235" "HSPC_241" "HSPC_247"
[487] "LT-HSC_061" "HSPC_211" "HSPC_218" "HSPC_224" "HSPC_230" "HSPC_236" "HSPC_242" "HSPC_248" "LT-HSC_062" "HSPC_212" "HSPC_219" "HSPC_225" "HSPC_231" "HSPC_237" "HSPC_243" "HSPC_249" "LT-HSC_063" "HSPC_213"
[505] "HSPC_220" "HSPC_226" "HSPC_232" "HSPC_238" "HSPC_244" "HSPC_250" "LT-HSC_064" "HSPC_214" "HSPC_221" "HSPC_227" "HSPC_233" "HSPC_239" "HSPC_245" "HSPC_251" "LT-HSC_065" "HSPC_215" "HSPC_222" "HSPC_228"
[523] "HSPC_234" "HSPC_240" "HSPC_246" "HSPC_252" "LT-HSC_066" "HSPC_216" "Prog_217" "Prog_223" "Prog_229" "Prog_235" "Prog_241" "Prog_247" "LT-HSC_067" "Prog_211" "Prog_218" "Prog_224" "Prog_230" "Prog_236"
[541] "Prog_242" "Prog_248" "LT-HSC_068" "Prog_212" "Prog_219" "Prog_225" "Prog_231" "Prog_237" "Prog_243" "Prog_249" "LT-HSC_069" "Prog_213" "Prog_220" "Prog_226" "Prog_232" "Prog_238" "Prog_244" "Prog_250"
[559] "LT-HSC_070" "Prog_214" "Prog_221" "Prog_227" "Prog_233" "Prog_239" "Prog_245" "Prog_251" "LT-HSC_071" "Prog_215" "Prog_222" "Prog_228" "Prog_234" "Prog_240" "Prog_246" "Prog_252" "LT-HSC_072" "Prog_216"
[577] "HSPC_259" "HSPC_265" "HSPC_271" "HSPC_277" "HSPC_283" "HSPC_289" "LT-HSC_073" "HSPC_253" "HSPC_260" "HSPC_266" "HSPC_272" "HSPC_278" "HSPC_284" "HSPC_290" "LT-HSC_074" "HSPC_254" "HSPC_261" "HSPC_267"
[595] "HSPC_273" "HSPC_279" "HSPC_285" "HSPC_291" "LT-HSC_075" "HSPC_255" "HSPC_262" "HSPC_268" "HSPC_274" "HSPC_280" "HSPC_286" "HSPC_292" "LT-HSC_076" "HSPC_256" "HSPC_263" "HSPC_269" "HSPC_275" "HSPC_281"
[613] "HSPC_287" "HSPC_293" "LT-HSC_077" "HSPC_257" "HSPC_264" "HSPC_270" "HSPC_276" "HSPC_282" "HSPC_288" "HSPC_294" "LT-HSC_078" "HSPC_258" "Prog_259" "Prog_265" "Prog_271" "Prog_277" "Prog_283" "Prog_289"
[631] "LT-HSC_079" "Prog_253" "Prog_260" "Prog_266" "Prog_272" "Prog_278" "Prog_284" "Prog_290" "LT-HSC_080" "Prog_254" "Prog_261" "Prog_267" "Prog_273" "Prog_279" "Prog_285" "Prog_291" "LT-HSC_081" "Prog_255"
[649] "Prog_262" "Prog_268" "Prog_274" "Prog_280" "Prog_286" "Prog_292" "LT-HSC_082" "Prog_256" "Prog_263" "Prog_269" "Prog_275" "Prog_281" "Prog_287" "Prog_293" "LT-HSC_083" "Prog_257" "Prog_264" "Prog_270"
[667] "Prog_276" "Prog_282" "Prog_288" "Prog_294" "LT-HSC_084" "Prog_258" "HSPC_301" "HSPC_307" "HSPC_313" "HSPC_319" "HSPC_325" "HSPC_331" "LT-HSC_085" "HSPC_295" "HSPC_302" "HSPC_308" "HSPC_314" "HSPC_320"
[685] "HSPC_326" "HSPC_332" "LT-HSC_086" "HSPC_296" "HSPC_303" "HSPC_309" "HSPC_315" "HSPC_321" "HSPC_327" "HSPC_333" "LT-HSC_087" "HSPC_297" "HSPC_304" "HSPC_310" "HSPC_316" "HSPC_322" "HSPC_328" "HSPC_334"
[703] "LT-HSC_088" "HSPC_298" "HSPC_305" "HSPC_311" "HSPC_317" "HSPC_323" "HSPC_329" "HSPC_335" "LT-HSC_089" "HSPC_299" "HSPC_306" "HSPC_312" "HSPC_318" "HSPC_324" "HSPC_330" "HSPC_336" "LT-HSC_090" "HSPC_300"
[721] "Prog_301" "Prog_307" "Prog_313" "Prog_319" "Prog_325" "Prog_331" "LT-HSC_091" "Prog_295" "Prog_302" "Prog_308" "Prog_314" "Prog_320" "Prog_326" "Prog_332" "LT-HSC_092" "Prog_296" "Prog_303" "Prog_309"
[739] "Prog_315" "Prog_321" "Prog_327" "Prog_333" "LT-HSC_093" "Prog_297" "Prog_304" "Prog_310" "Prog_316" "Prog_322" "Prog_328" "Prog_334" "LT-HSC_094" "Prog_298" "Prog_305" "Prog_311" "Prog_317" "Prog_323"
[757] "Prog_329" "Prog_335" "LT-HSC_095" "Prog_299" "Prog_306" "Prog_312" "Prog_318" "Prog_324" "Prog_330" "Prog_336" "LT-HSC_096" "Prog_300" "HSPC_343" "HSPC_349" "HSPC_355" "HSPC_361" "HSPC_367" "HSPC_373"
[775] "LT-HSC_097" "HSPC_337" "HSPC_344" "HSPC_350" "HSPC_356" "HSPC_362" "HSPC_368" "HSPC_374" "LT-HSC_098" "HSPC_338" "HSPC_345" "HSPC_351" "HSPC_357" "HSPC_363" "HSPC_369" "HSPC_375" "LT-HSC_099" "HSPC_339"
[793] "HSPC_346" "HSPC_352" "HSPC_358" "HSPC_364" "HSPC_370" "HSPC_376" "LT-HSC_100" "HSPC_340" "HSPC_347" "HSPC_353" "HSPC_359" "HSPC_365" "HSPC_371" "HSPC_377" "LT-HSC_101" "HSPC_341" "HSPC_348" "HSPC_354"
[811] "HSPC_360" "HSPC_366" "HSPC_372" "HSPC_378" "LT-HSC_102" "HSPC_342" "Prog_343" "Prog_349" "Prog_355" "Prog_361" "Prog_367" "Prog_373" "LT-HSC_103" "Prog_337" "Prog_344" "Prog_350" "Prog_356" "Prog_362"
[829] "Prog_368" "Prog_374" "LT-HSC_104" "Prog_338" "Prog_345" "Prog_351" "Prog_357" "Prog_363" "Prog_369" "Prog_375" "LT-HSC_105" "Prog_339" "Prog_346" "Prog_352" "Prog_358" "Prog_364" "Prog_370" "Prog_376"
[847] "LT-HSC_106" "Prog_340" "Prog_347" "Prog_353" "Prog_359" "Prog_365" "Prog_371" "Prog_377" "LT-HSC_107" "Prog_341" "Prog_348" "Prog_354" "Prog_360" "Prog_366" "Prog_372" "Prog_378" "LT-HSC_108" "Prog_342"
[865] "HSPC_385" "HSPC_391" "HSPC_397" "HSPC_403" "HSPC_409" "HSPC_415" "HSPC_421" "HSPC_379" "HSPC_386" "HSPC_392" "HSPC_398" "HSPC_404" "HSPC_410" "HSPC_416" "HSPC_422" "HSPC_380" "HSPC_387" "HSPC_393"
[883] "HSPC_399" "HSPC_405" "HSPC_411" "HSPC_417" "HSPC_423" "HSPC_381" "HSPC_388" "HSPC_394" "HSPC_400" "HSPC_406" "HSPC_412" "HSPC_418" "HSPC_424" "HSPC_382" "HSPC_389" "HSPC_395" "HSPC_401" "HSPC_407"
[901] "HSPC_413" "HSPC_419" "HSPC_425" "HSPC_383" "HSPC_390" "HSPC_396" "HSPC_402" "HSPC_408" "HSPC_414" "HSPC_420" "HSPC_426" "HSPC_384" "Prog_385" "Prog_391" "Prog_397" "Prog_403" "Prog_409" "Prog_415"
[919] "Prog_421" "Prog_379" "Prog_386" "Prog_392" "Prog_398" "Prog_404" "Prog_410" "Prog_416" "Prog_422" "Prog_380" "Prog_387" "Prog_393" "Prog_399" "Prog_405" "Prog_411" "Prog_417" "Prog_423" "Prog_381"
[937] "Prog_388" "Prog_394" "Prog_400" "Prog_406" "Prog_412" "Prog_418" "Prog_424" "Prog_382" "Prog_389" "Prog_395" "Prog_401" "Prog_407" "Prog_413" "Prog_419" "Prog_425" "Prog_383" "Prog_390" "Prog_396"
[955] "Prog_402" "Prog_408" "Prog_414" "Prog_420" "Prog_426" "Prog_384" "HSPC_433" "HSPC_439" "HSPC_445" "HSPC_451" "HSPC_457" "HSPC_463" "LT-HSC_109" "HSPC_427" "HSPC_434" "HSPC_440" "HSPC_446" "HSPC_452"
[973] "HSPC_458" "HSPC_464" "LT-HSC_110" "HSPC_428" "HSPC_435" "HSPC_441" "HSPC_447" "HSPC_453" "HSPC_459" "HSPC_465" "LT-HSC_111" "HSPC_429" "HSPC_436" "HSPC_442" "HSPC_448" "HSPC_454" "HSPC_460" "HSPC_466"
[991] "LT-HSC_112" "HSPC_430" "HSPC_437" "HSPC_443" "HSPC_449" "HSPC_455" "HSPC_461" "HSPC_467" "LT-HSC_113" "HSPC_431"
[ reached 'max' / getOption("max.print") -- omitted 920 entries ]ncol(sce_seurat)[1] 1920nrow(sce_seurat)[1] 500[1] "HSPC_007" "HSPC_013" "HSPC_019" "HSPC_025" "HSPC_031" "HSPC_037"[1] "ENSMUSG00000076609" "ENSMUSG00000021250" "ENSMUSG00000076617" "ENSMUSG00000075602" "ENSMUSG00000006389" "ENSMUSG00000041481"# cell data access
head(sce_seurat[[]]) orig.ident nCount_endogenous nFeature_endogenous gate broad broad.mpp fine fine.mpp ESLAM HSC1 projected metrics.X__no_feature metrics.X__ambiguous metrics.X__too_low_aQual metrics.X__not_aligned metrics.X__alignment_not_unique
HSPC_007 HSPC 1262 381 HSPC <NA> <NA> <NA> <NA> FALSE FALSE FALSE 194829 5022 0 5820455 0
HSPC_013 HSPC 5906 409 HSPC LMPP <NA> LMPP <NA> FALSE FALSE FALSE 110530 15271 0 1562724 0
HSPC_019 HSPC 1002 355 HSPC LMPP <NA> <NA> <NA> FALSE FALSE FALSE 86825 2708 0 1407254 0
HSPC_025 HSPC 58177 434 HSPC MPP MPP1 <NA> <NA> FALSE FALSE FALSE 212206 107278 0 1810368 0
HSPC_031 HSPC 118570 435 HSPC MPP STHSC <NA> <NA> FALSE FALSE FALSE 690411 227480 0 6097116 0
HSPC_037 HSPC 54029 445 HSPC MPP STHSC <NA> <NA> FALSE FALSE FALSE 242472 126874 0 2267894 0
label sizeFactor
HSPC_007 HSPC 0.02724693
HSPC_013 HSPC 0.09042153
HSPC_019 HSPC 0.01992108
HSPC_025 HSPC 0.52179199
HSPC_031 HSPC 1.10623062
HSPC_037 HSPC 0.51431538head(sce_seurat$label)HSPC_007 HSPC_013 HSPC_019 HSPC_025 HSPC_031 HSPC_037
"HSPC" "HSPC" "HSPC" "HSPC" "HSPC" "HSPC" # Counts access
Seurat::GetAssayData(object = sce_seurat, layer = "counts")[1:10, 1:10]10 x 10 sparse Matrix of class "dgCMatrix" [[ suppressing 10 column names 'HSPC_007', 'HSPC_013', 'HSPC_019' ... ]]
ENSMUSG00000076609 30 16 7 17 19 11 1359 13 15 6
ENSMUSG00000021250 3 2 4 . 1 3 118 2 69 41
ENSMUSG00000076617 40 54 13 29 298 417 1107 9 16 15
ENSMUSG00000075602 3 248 7 537 640 7 300 1530 324 971
ENSMUSG00000006389 3 4 1 1171 6 2 271 5497 293 192
ENSMUSG00000041481 3 . 1 3 1 3 177 5 1 192
ENSMUSG00000024190 1 29 1 1733 1 5 20 3 1 1
ENSMUSG00000003949 4 15 3 175 915 1131 41 1465 330 258
ENSMUSG00000052684 1 4 2 144 82 5 142 578 . 78
ENSMUSG00000026358 . 27 6 344 3 9 3 3612 185 .Seurat::GetAssayData(object = sce_seurat, layer = "data")[1:10, 1:10]10 x 10 sparse Matrix of class "dgCMatrix" [[ suppressing 10 column names 'HSPC_007', 'HSPC_013', 'HSPC_019' ... ]]
ENSMUSG00000076609 10.105963 7.475320 8.461015 5.069532 4.1839186 4.484632 13.057990 2.7927556 4.907536 4.256554
ENSMUSG00000021250 6.795769 4.530984 7.656727 . 0.9290112 2.772518 9.534081 0.9352591 7.071052 6.963227
ENSMUSG00000076617 10.520673 9.224491 9.352209 5.822162 8.0788620 9.664957 12.762138 2.3519259 4.997638 5.532469
ENSMUSG00000075602 6.795769 11.421912 8.461015 10.008633 9.1787674 3.868916 10.879076 9.4488576 9.293908 11.517886
ENSMUSG00000006389 6.795769 5.499439 5.678018 11.132621 2.6834323 2.289440 10.732488 11.2924799 9.149058 9.181516
ENSMUSG00000041481 6.795769 . 5.678018 2.754763 0.9290112 2.772518 10.118395 1.7139531 1.552981 9.181516
ENSMUSG00000024190 5.236545 8.329662 5.678018 11.697944 0.9290112 3.422457 6.982870 1.2438829 1.552981 2.006833
ENSMUSG00000003949 7.207556 7.382751 7.244071 8.393960 9.6937189 11.103314 8.012640 9.3863182 9.320338 9.607145
ENSMUSG00000052684 5.236545 5.499439 6.663859 8.113597 6.2312326 3.422457 9.800857 8.0478635 . 7.885582
ENSMUSG00000026358 . 8.226901 8.239305 9.366905 1.8921625 4.209375 4.309131 10.6869334 8.487165 . # Embeddings
head(Seurat::Embeddings(object = sce_seurat, reduction = "diffusion")) DC_1 DC_2 DC_3
HSPC_007 NA NA NA
HSPC_013 NA NA NA
HSPC_019 NA NA NA
HSPC_025 -0.01101623 -0.001401563 0.01644595
HSPC_031 -0.01378359 -0.011040897 0.01367213
HSPC_037 -0.01377982 -0.003303141 0.03332661Try to load the raw 10X single-cell RNA-seq data downloaded yesterday (from Lier et al.) into a SingleCellExperiment object using DropletUtils package
R
library(SingleCellExperiment)
sce <- DropletUtils::read10xCounts('~/Share/data_wrangling/counts/outs/filtered_feature_bc_matrix.h5')
sceclass: SingleCellExperiment
dim: 32285 686
metadata(1): Samples
assays(1): counts
rownames(32285): ENSMUSG00000051951 ENSMUSG00000089699 ... ENSMUSG00000095019 ENSMUSG00000095041
rowData names(3): ID Symbol Type
colnames: NULL
colData names(2): Sample BarcodeFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'reducedDimNames(0):
mainExpName: NULLFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'altExpNames(0):colData(sce)DataFrame with 686 rows and 2 columns
Sample Barcode
<character> <character>
1 ~/Share/data_wrangli.. AAACCTGAGGGTCTCC-1
2 ~/Share/data_wrangli.. AAACCTGCACGGCGTT-1
3 ~/Share/data_wrangli.. AAACGGGTCTGCAAGT-1
4 ~/Share/data_wrangli.. AAAGATGGTTCGAATC-1
5 ~/Share/data_wrangli.. AAAGCAAAGGTGGGTT-1
... ... ...
682 ~/Share/data_wrangli.. TTTCCTCAGACTCGGA-1
683 ~/Share/data_wrangli.. TTTCCTCTCGGAGGTA-1
684 ~/Share/data_wrangli.. TTTGTCAAGTGTCCCG-1
685 ~/Share/data_wrangli.. TTTGTCACATGGAATA-1
686 ~/Share/data_wrangli.. TTTGTCAGTCGAGATG-1rowData(sce)DataFrame with 32285 rows and 3 columns
ID Symbol Type
<character> <character> <character>
ENSMUSG00000051951 ENSMUSG00000051951 Xkr4 Gene Expression
ENSMUSG00000089699 ENSMUSG00000089699 Gm1992 Gene Expression
ENSMUSG00000102331 ENSMUSG00000102331 Gm19938 Gene Expression
ENSMUSG00000102343 ENSMUSG00000102343 Gm37381 Gene Expression
ENSMUSG00000025900 ENSMUSG00000025900 Rp1 Gene Expression
... ... ... ...
ENSMUSG00000095523 ENSMUSG00000095523 AC124606.1 Gene Expression
ENSMUSG00000095475 ENSMUSG00000095475 AC133095.2 Gene Expression
ENSMUSG00000094855 ENSMUSG00000094855 AC133095.1 Gene Expression
ENSMUSG00000095019 ENSMUSG00000095019 AC234645.1 Gene Expression
ENSMUSG00000095041 ENSMUSG00000095041 AC149090.1 Gene ExpressionPublic single-cell RNA-seq data can be retrieved from within R directly, thanks to several data packages, for instance scRNAseq or HCAData.
Check out the He et al., Genome Biol. 2020 paper. Can you find a way to load the scRNAseq data from this paper without having to leave the R console?
R
organs <- scRNAseq::HeOrganAtlasData(ensembl = TRUE)Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'loading from cacheWarning: Unable to map 1056 of 12021 requested IDs.Found more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'Also defined by 'SeuratObject'organsclass: SingleCellExperiment
dim: 10965 84363
metadata(0):
assays(1): counts
rownames(10965): ENSG00000225880 ENSG00000188976 ... ENSG00000273748 ENSG00000271254
rowData names(1): originalName
colnames(84363): AAACCTGAGACACTAA-1 AAACCTGAGCATGGCA-1 ... TTTGTCATCTCTAGGA-1 TTTGTCATCTGCGACG-1
colData names(10): Tissue nCount_RNA ... reclustered.broad reclustered.fineFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'reducedDimNames(1): TSNE
mainExpName: NULLFound more than one class "package_version" in cache; using the first, from namespace 'alabaster.base'
Also defined by 'SeuratObject'altExpNames(0):The interest of this approach is that one can recover a full-fledged SingleCellExperiment (often) provided by the authors of the corresponding study. This means that lots of information, such as batch ID, clustering, cell annotation, etc., may be readily available.
Check the data available for cells/features in the dataset from He et al..
R
colData(organs)DataFrame with 84363 rows and 10 columns
Tissue nCount_RNA nFeature_RNA percent.mito RNA_snn_res.orig seurat_clusters Cell_type_in_each_tissue Cell_type_in_merged_data reclustered.broad reclustered.fine
<character> <integer> <integer> <numeric> <integer> <integer> <character> <character> <character> <character>
AAACCTGAGACACTAA-1 Bladder 1152 610 0.0789931 7 15 Monocyte Bladder Monocyte Myeloid Classical_Mon_AQP9
AAACCTGAGCATGGCA-1 Bladder 3551 1415 0.0568854 14 8 Macrophage HLA-DPB1_.. Macrophage C1QB Myeloid cDC2
AAACCTGAGCTGAACG-1 Bladder 1842 632 0.0266015 4 1 T Cell IL7R_high Bla.. T Cell IL7R CD4 TCM_KLF2
AAACCTGCAAGCTGGA-1 Bladder 1599 890 0.0268918 13 13 Fibroblast APOD_high.. Fibroblast PTGDS FibSmo Fib_CHI3L1_high
AAACCTGCATTACCTT-1 Bladder 1347 613 0.0541945 1 2 T Cell CCL5_high Bla.. T Cell XCL1 NA NA
... ... ... ... ... ... ... ... ... ... ...
TTTGTCATCAGTTGAC-1 Trachea 3334 1461 0.0422915 13 36 B Cell Trachea B Cell MS4A1 B_and_plasma Memory_B_TNF
TTTGTCATCATCATTC-1 Trachea 6634 2271 0.0611999 7 8 Macrophage/Monocyte .. Macrophage C1QB Myeloid Mac_SPP1
TTTGTCATCTACGAGT-1 Trachea 2255 769 0.2119734 13 2 B Cell Trachea T Cell XCL1 B_and_plasma Plasma_IGKV3-20
TTTGTCATCTCTAGGA-1 Trachea 1568 832 0.0554847 0 21 CD8 T Cell CCL5_high.. T Cell CCL5 CD8 TRM_PRR4
TTTGTCATCTGCGACG-1 Trachea 1907 999 0.0167803 18 20 NK/T Cell Trachea NK/T Cell GNLY NA NAtable(organs$Tissue)
Bladder Blood Common.bile.duct Esophagus Heart Liver Lymph.node Marrow Muscle Rectum Skin Small.intestine Spleen Stomach
7572 1407 3160 9117 7881 2839 7771 3230 5732 6280 7710 4312 4512 5318
Trachea
7522 table(organs$reclustered.fine, organs$Tissue)
Bladder Blood Common.bile.duct Esophagus Heart Liver Lymph.node Marrow Muscle Rectum Skin Small.intestine Spleen Stomach Trachea
Absorptive Cell 0 0 0 0 0 0 0 0 0 0 0 170 0 0 0
Basal Epithelial Cell APOE_high 4 0 0 3 0 0 0 0 0 0 504 1 0 0 1
Basal Epithelial Cell IFITM1_high 0 0 1 2 0 0 0 0 0 0 0 0 0 0 441
Basal Epithelial Cell MMP10_high 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1747
Basal Epithelial Cell POSTN_high 0 0 0 0 0 0 0 0 0 0 545 0 0 0 0
Basal Epithelial Cell SERPINB3_high 0 0 0 0 0 0 0 0 0 0 0 0 0 0 839
BEC_ACKR1 105 0 2 177 589 2 0 0 179 38 60 0 0 3 55
BEC_APOC1 0 0 0 0 3 2 0 0 49 0 0 0 0 0 0
BEC_CA4 13 0 1 13 824 9 0 0 53 6 3 0 0 1 7
BEC_CTSC 38 0 0 66 38 1 0 0 12 1 1058 0 0 0 70
BEC_FABP4 3 0 0 1 22 0 0 0 740 1 1 0 0 0 2
BEC_IGFBP3 19 0 13 63 201 27 0 0 41 18 245 0 0 7 56
BEC_PHLDA1 80 0 6 449 36 1 0 0 2 3 56 0 0 1 29
BEC_PRSS23 5 0 74 0 0 3 0 0 0 2 2 0 0 1 5
BEC_RTEL1-TNFRSF6B 15 0 2 25 14 3 0 0 3 10 11 0 0 7 642
BEC_TIMP1 0 0 0 0 0 157 0 0 0 0 0 0 0 1 0
BEC_TNFRSF4 0 0 0 4 22 3 0 0 65 0 3 0 0 0 1
cDC1 36 0 4 1 1 0 12 0 0 2 1 0 1 0 7
cDC2 122 0 12 121 117 3 0 2 4 62 13 1 3 5 18
Cholangiocyte FXYD2_high 0 0 625 0 0 6 0 0 0 0 0 0 0 0 0
Cholangiocyte HIST1H2AM_high 0 0 83 0 0 0 0 0 0 0 0 0 0 0 0
Classical_Mon_AQP9 217 0 22 11 21 277 6 1 24 38 13 3 91 37 106
Classical_Mon_HCAR3 13 0 307 2 3 23 3 0 0 4 3 0 0 2 33
Classical_Mon_S100A12 26 1 12 3 9 130 1 0 7 3 9 0 81 21 11
Classical_Mon_S100A8 11 69 67 0 1 86 0 45 1 3 3 0 9 2 14
Enterocyte APOA1_high 0 0 0 0 0 0 0 0 0 0 0 654 0 0 3
Enterocyte APOB_high 0 0 0 0 0 0 0 0 0 0 0 586 0 0 4
Enterocyte PCK1_high 0 0 0 0 0 0 0 0 0 0 0 665 0 0 4
Enterocyte PRAP1_high 0 0 0 0 0 0 0 0 0 0 0 556 0 0 1
Enterocyte RBP2_high 0 0 0 0 0 0 0 0 0 0 0 412 0 0 1
Epithelial Cell DCD_high 1 0 2 41 0 0 0 0 0 0 73 0 0 1 3
Epithelial Cell FABP4_high 116 0 1 0 0 0 0 0 0 0 0 0 0 7 5
Fib_ANGPTL7_high 82 0 30 86 31 0 0 0 4 2 12 0 0 0 10
Fib_APOD_high 0 0 0 4 0 0 0 0 850 0 0 0 0 0 0
Fib_C1QTNF3_high 7 0 1 1291 2 0 0 0 5 3 2 0 0 4 0
Fib_CHI3L1_high 167 0 40 76 6 0 0 0 1 44 0 0 0 9 329
Fib_CRABP1_high 0 0 0 1 0 0 0 0 0 0 0 0 0 0 281
Fib_CXCL14_high 11 0 0 4 4 0 0 0 4 3 687 0 0 0 4
Fib_IGFBP3_high 635 0 0 2 0 0 0 0 0 6 0 0 0 2 0
Fib_IGFBP6_high 2 0 0 573 0 0 0 0 0 2 0 0 0 8 0
Fib_MT_high 22 0 1 74 4 0 0 0 0 21 8 0 0 10 13
Fib_PCOLCE2_high 4 0 0 0 1251 0 0 0 1 2 2 0 0 3 6
Fib_PTGDS_high 1 0 2 781 1 0 0 0 0 2 0 0 0 0 1
Fib_PTN_high 1 0 0 10 2467 1 0 0 1 4 6 0 0 2 7
Fib_SFRP2_high 5 0 0 0 0 0 0 0 0 328 0 0 0 10 0
Fib_SLIT3_high 1110 0 7 137 20 0 0 0 0 16 2 0 0 23 3
FibSmo_ADAMDEC1_high 1 0 0 1 0 0 0 0 0 80 0 0 0 30 1
FibSmo_FOXF1_high 199 0 0 0 0 0 0 0 0 0 0 0 0 0 0
FibSmo_LINC01082_high 922 0 0 0 0 0 0 0 0 10 0 0 0 2 0
FibSmo_SLC14A1_high 841 0 1 0 0 0 0 0 0 0 0 0 0 1 0
FibSmo_THBS4_high 1 0 0 2 0 0 0 0 0 731 0 0 0 4 1
Follicular Epithelial Cell 0 0 0 1 0 0 0 0 0 0 645 0 0 0 0
Goblet Cell 0 0 0 0 0 0 0 0 0 205 0 47 0 9 22
Granular Epithelial Cell 0 0 0 0 0 0 0 0 0 0 224 0 0 0 0
Hepatic Oval Cell 0 0 4 0 0 48 0 0 0 0 0 0 0 0 0
High Proliferation Epithelial Cell TOP2A_high 0 0 0 0 0 0 0 0 0 0 229 1 0 0 0
High Proliferation Epithelial Cell UBE2C_high 0 0 1 416 0 0 0 0 0 0 6 0 0 0 2
IEL_TMIGD2 0 0 0 0 0 0 0 0 0 4 0 1 0 443 0
IEL_TRBV7-3 0 0 1 0 0 0 0 0 0 0 0 0 0 0 243
Intermediate_Mon_CCL20 0 0 0 0 0 0 0 0 0 232 0 0 0 1 3
Intermediate_Mon_FN1 33 0 2 1 9 7 0 0 4 432 0 4 1 4 50
Langerhans 0 0 0 0 0 0 0 0 0 0 72 0 0 0 0
LEC_CCL21 14 0 0 48 3 13 0 0 0 5 26 0 0 0 13
LEC_FCN3 0 0 0 0 0 129 0 0 0 0 0 0 0 0 0
Mac_APOE 5 0 59 0 0 3 0 0 0 0 1 0 0 1 5
Mac_FTL 15 2 4 2 3 5 0 2 1 1 0 2 4 0 5
[ reached 'max' / getOption("max.print") -- omitted 66 rows ]To compare the two different approaches, try preparing both a SingleCellExperiment or a Seurat object from scratch, using the matrix files generated in the previous lab. Read the documentation of the two related packages to understand how to do this.
This will be extensively covered in the next lab for everybody.