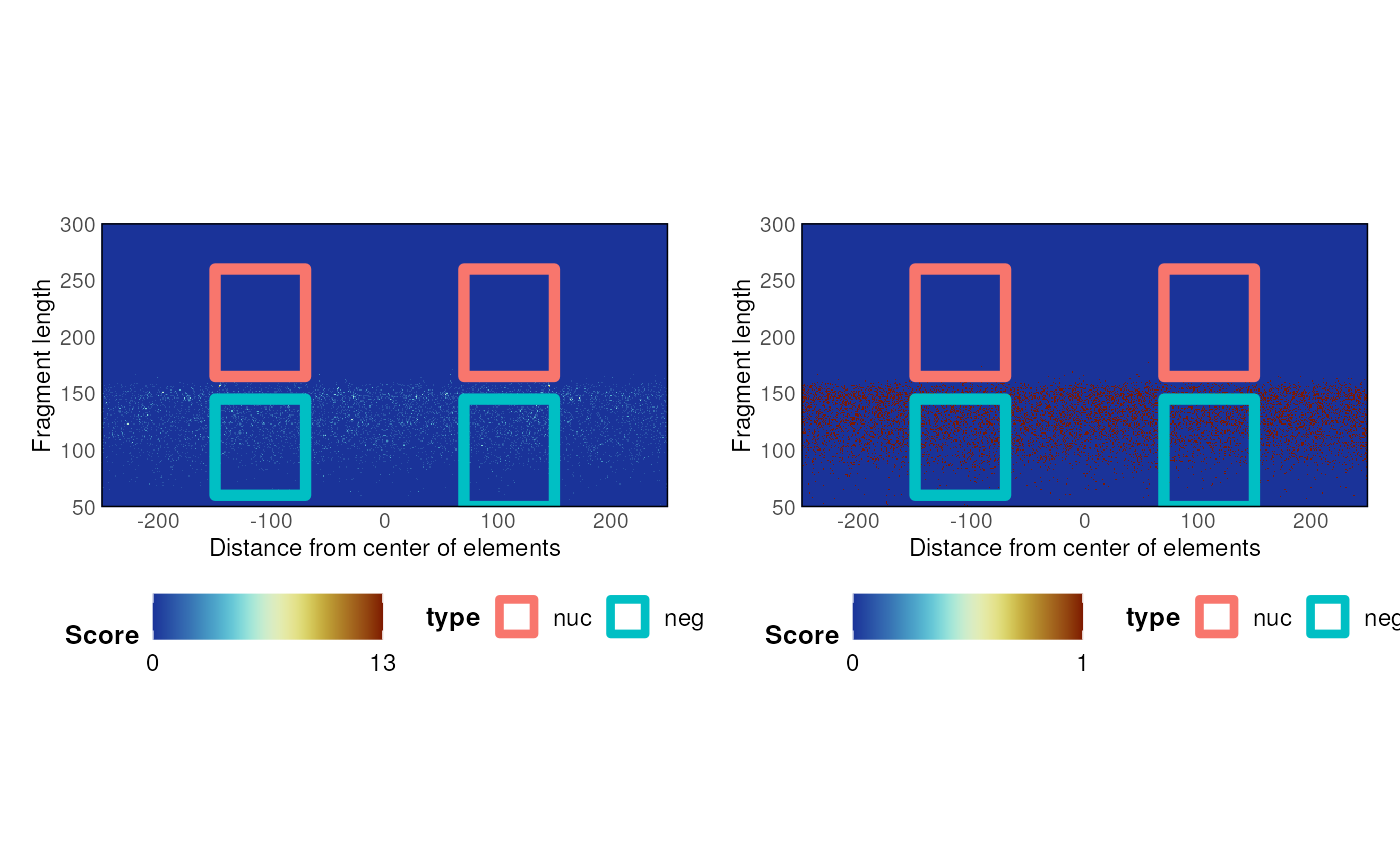
A function to compute nucleosome enrichment over a set of GRanges
Source:R/nucleosome_enrichment.R
nucleosomeEnrichment.GRanges.RdA function to compute nucleosome enrichment over a set of GRanges
# S3 method for GRanges
nucleosomeEnrichment(x, granges, plus1_nuc_only = FALSE, verbose = TRUE, ...)Arguments
- x
GRanges, paired-end fragments
- granges
GRanges, loci to map the fragments onto
- plus1_nuc_only
Boolean, should compute nucleosome enrichment only for +1 nucleosome?
- verbose
Boolean
- ...
additional parameters
Value
list
Examples
data(bam_test)
data(ce11_proms)
n <- nucleosomeEnrichment(bam_test, ce11_proms)
#> Computing Vmat...
#> Computing background...
#> Computing enrichment...
n$fisher_test
#>
#> Fisher's Exact Test for Count Data
#>
#> data: matrix(vec, ncol = 2)
#> p-value = 0.8227
#> alternative hypothesis: true odds ratio is not equal to 1
#> 95 percent confidence interval:
#> 0.4141458 3.0139383
#> sample estimates:
#> odds ratio
#> 1.133024
#>
n$plot