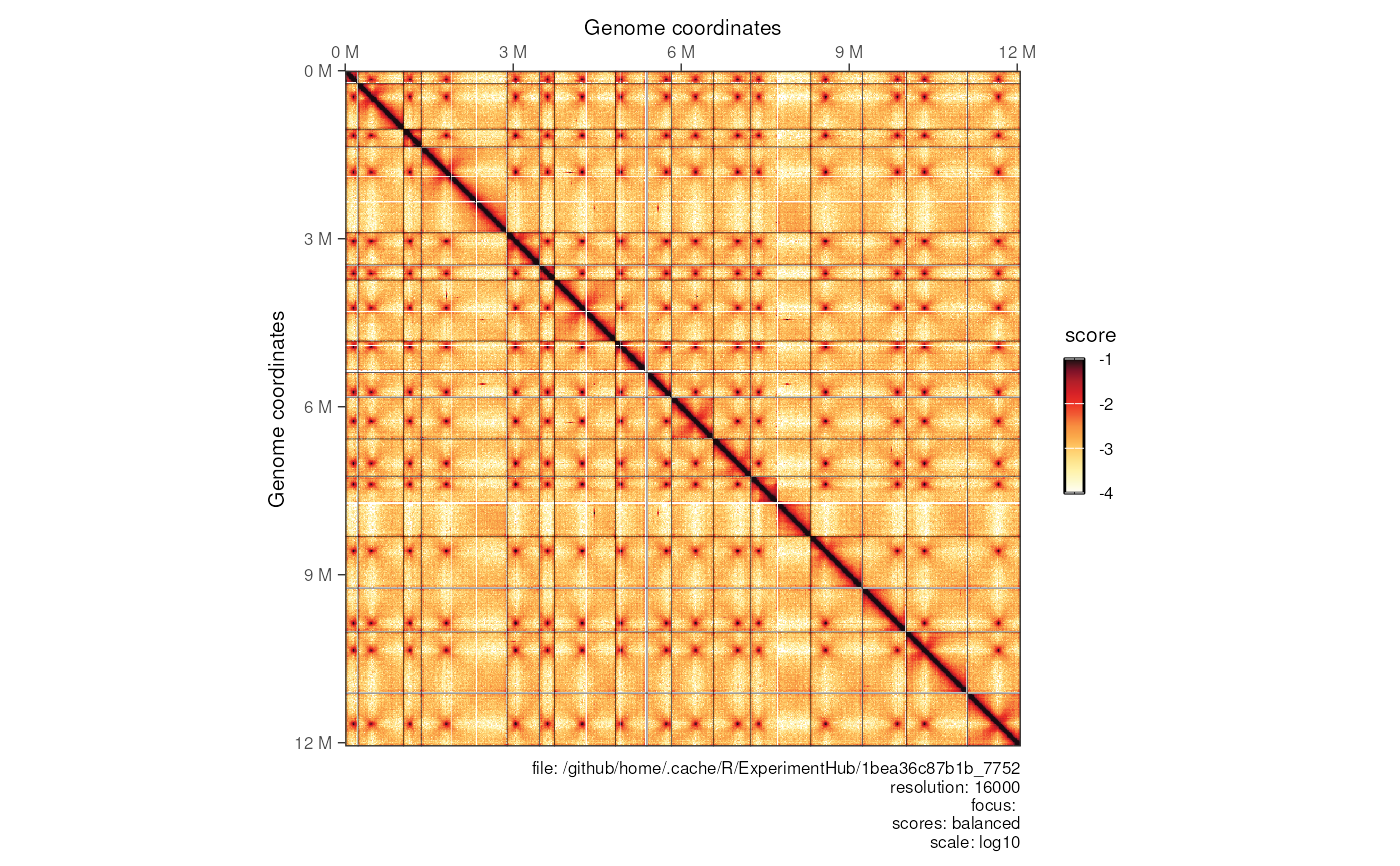
Plotting a contact matrix
plotMatrix(x, ...)
montage(x, ...)
# S4 method for class 'HiCExperiment'
plotMatrix(
x,
compare.to = NULL,
use.scores = "balanced",
scale = "log10",
maxDistance = NULL,
loops = NULL,
borders = NULL,
tracks = NULL,
limits = NULL,
dpi = 500,
rasterize = TRUE,
symmetrical = TRUE,
chrom_lines = TRUE,
show_grid = FALSE,
cmap = NULL,
caption = TRUE
)
# S4 method for class 'GInteractions'
plotMatrix(
x,
use.scores = NULL,
scale = "log10",
maxDistance = NULL,
loops = NULL,
borders = NULL,
tracks = NULL,
limits = NULL,
dpi = 500,
rasterize = TRUE,
symmetrical = TRUE,
chrom_lines = TRUE,
show_grid = FALSE,
cmap = NULL
)
# S4 method for class 'matrix'
plotMatrix(
x,
scale = "log10",
limits = NULL,
dpi = 500,
rasterize = TRUE,
cmap = NULL
)
# S4 method for class 'AggrHiCExperiment'
plotMatrix(
x,
use.scores = "balanced",
scale = "log10",
maxDistance = NULL,
loops = NULL,
borders = NULL,
limits = NULL,
dpi = 500,
rasterize = TRUE,
chrom_lines = TRUE,
show_grid = FALSE,
cmap = NULL,
caption = TRUE
)
# S4 method for class 'AggrHiCExperiment'
montage(
x,
use.scores = "balanced",
scale = "log10",
limits = NULL,
dpi = 500,
rasterize = TRUE,
cmap = NULL
)Arguments
- x
A HiCExperiment object
- ...
Extra arguments passed to the corresponding method.
- compare.to
Compare to a second HiC matrix in the lower left corner
- use.scores
Which scores to use in the heatmap
- scale
Any of 'log10', 'log2', 'linear', 'exp0.2' (Default: 'log10')
- maxDistance
maximum distance. If provided, the heatmap is plotted horizontally
- loops
Loops to plot on top of the heatmap, provided as
GInteractions- borders
Borders to plot on top of the heatmap, provided as
GRanges- tracks
Named list of bigwig tracks imported as
Rle- limits
color map limits
- dpi
DPI to create the plot (Default: 500)
- rasterize
Whether the generated heatmap is rasterized or vectorized (Default: TRUE)
- symmetrical
Whether to enforce a symetrical heatmap (Default: TRUE)
- chrom_lines
Whether to display separating lines between chromosomes, should any be necessary (Default: TRUE)
- show_grid
Whether to display an underlying grid (Default: FALSE)
- cmap
Color scale to use. (Default: bgrColors() if limits are c(-1, 1) and coolerColors() otherwise)
- caption
Whether to display a caption (Default: TRUE)
Value
ggplot object
Examples
contacts_yeast <- contacts_yeast()
#> see ?HiContactsData and browseVignettes('HiContactsData') for documentation
#> loading from cache
plotMatrix(
contacts_yeast,
use.scores = 'balanced',
scale = 'log10',
limits = c(-4, -1)
)